Database-building method for amplicon sequencing
An amplicon and sequencing technology, applied in the field of molecular biology, can solve the problems of unfavorable sample sequencing library construction, heavy workload, waste of time, etc., to increase library construction time and cost, improve efficiency, and increase the possibility of introducing mutations. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
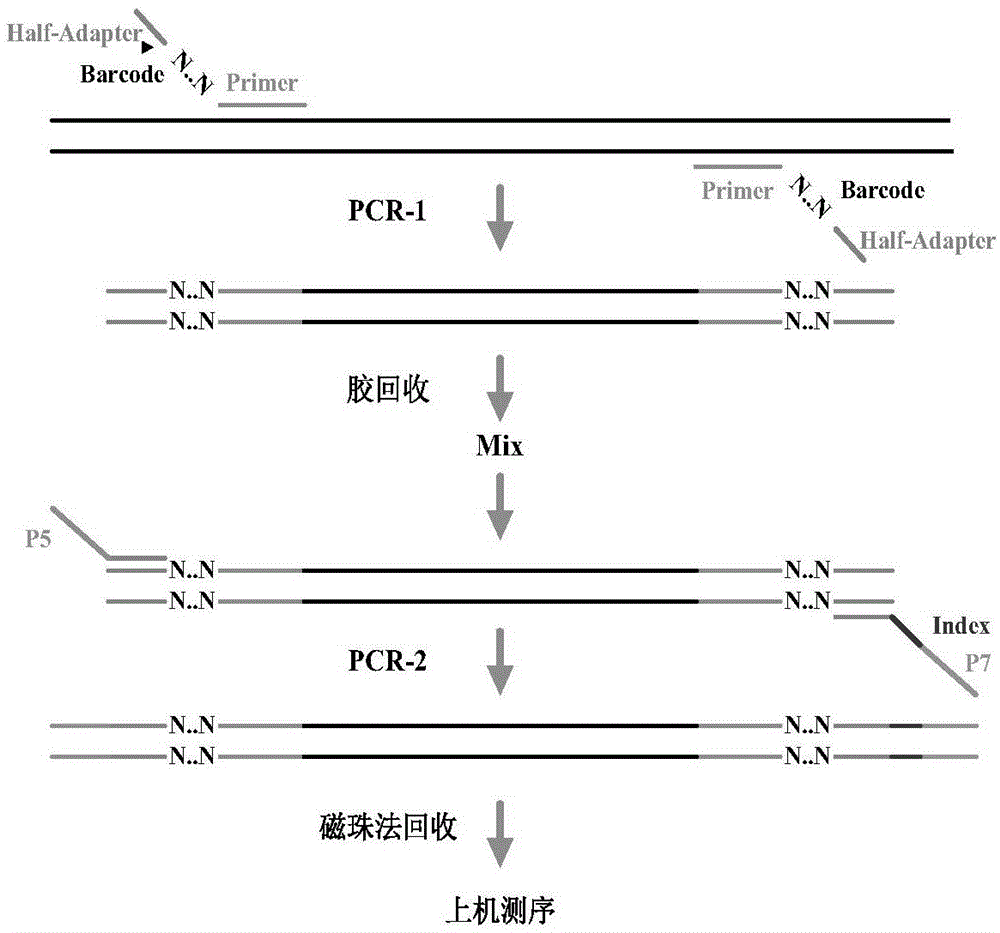
[0026] Firstly, the SOIL DNA KIT of Omega Company was used to extract the soil genomic DNA of 10 samples according to the conventional method, and the steps were omitted.
[0027] 1. PCR amplification of the enriched target region (PCR-1):
[0028] i. Primer Design
[0029] 1) The sequencing region is NF-NR;
[0030] 2) Design unique bases (N represents base sequence) and Adapter sequences at the 5' ends of primers NF and NR;
[0031] 3) The specific primer sequence information is shown in Table 1:
[0032] Forward primer NF: SEQ ID No 1;
[0033] Reverse primer NR: SEQ ID No 2.
[0034] Table 1 PCR-1 amplification primer sequences
[0035] Primer name
sequence (5'to3')
NF
CTTTCCCTACACGACGCTCTTCCGATCTNNNNNNAGAGTTTGATCCTGGCTCAG
NR
TGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNTTACCGCGGCTGCTGGCAC
[0036] ii. PCR amplification
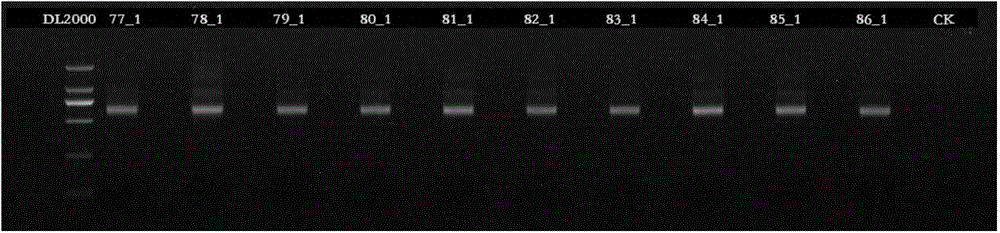
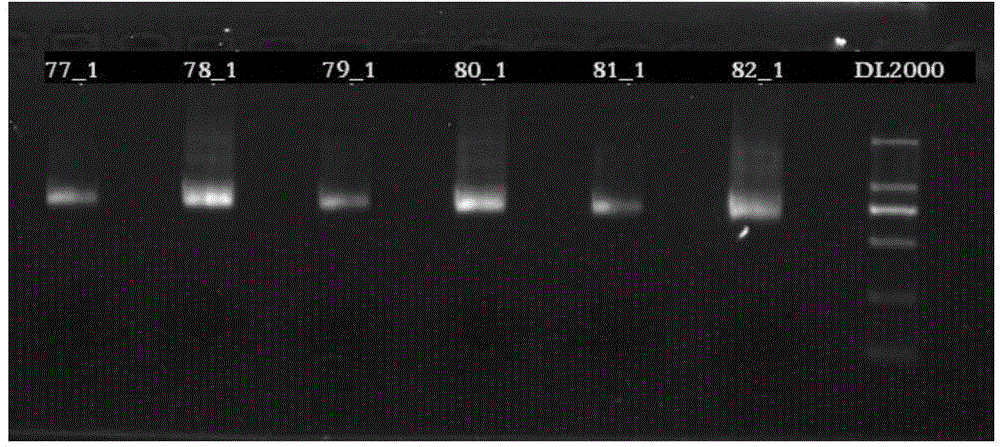
[0037] 1) PCR amplification is carried out to 10 sample genomic DNAs, and each sample is respectively PCR amplifie...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com