Modeling method of non-rodent animal model for studying intestinal flora
A modeling method, the technology of intestinal flora, applied in the field of intestinal symbiotic bacteria research, can solve the problems such as no successful reports, and achieve the effect of stable model, short time and low mortality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
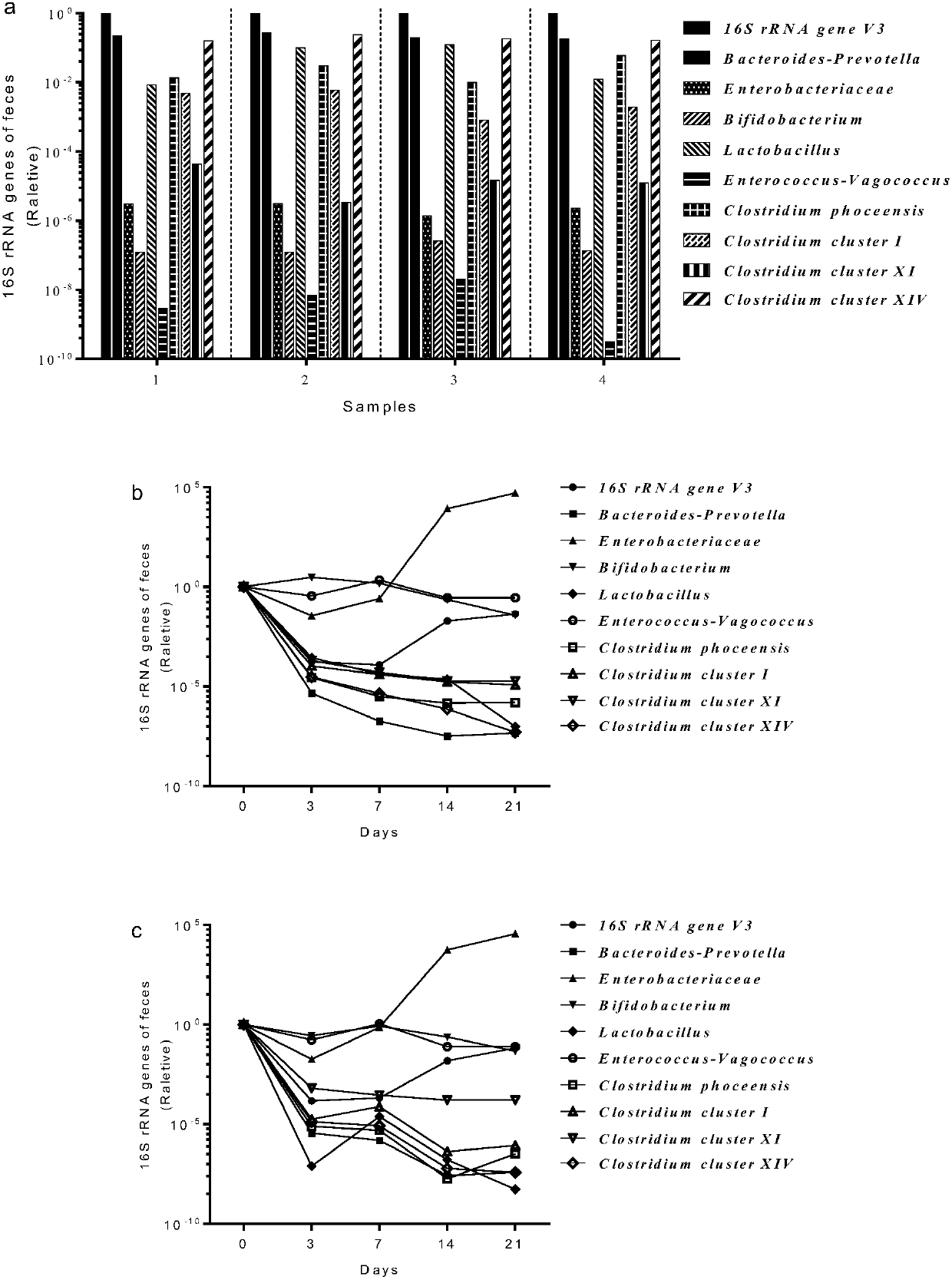
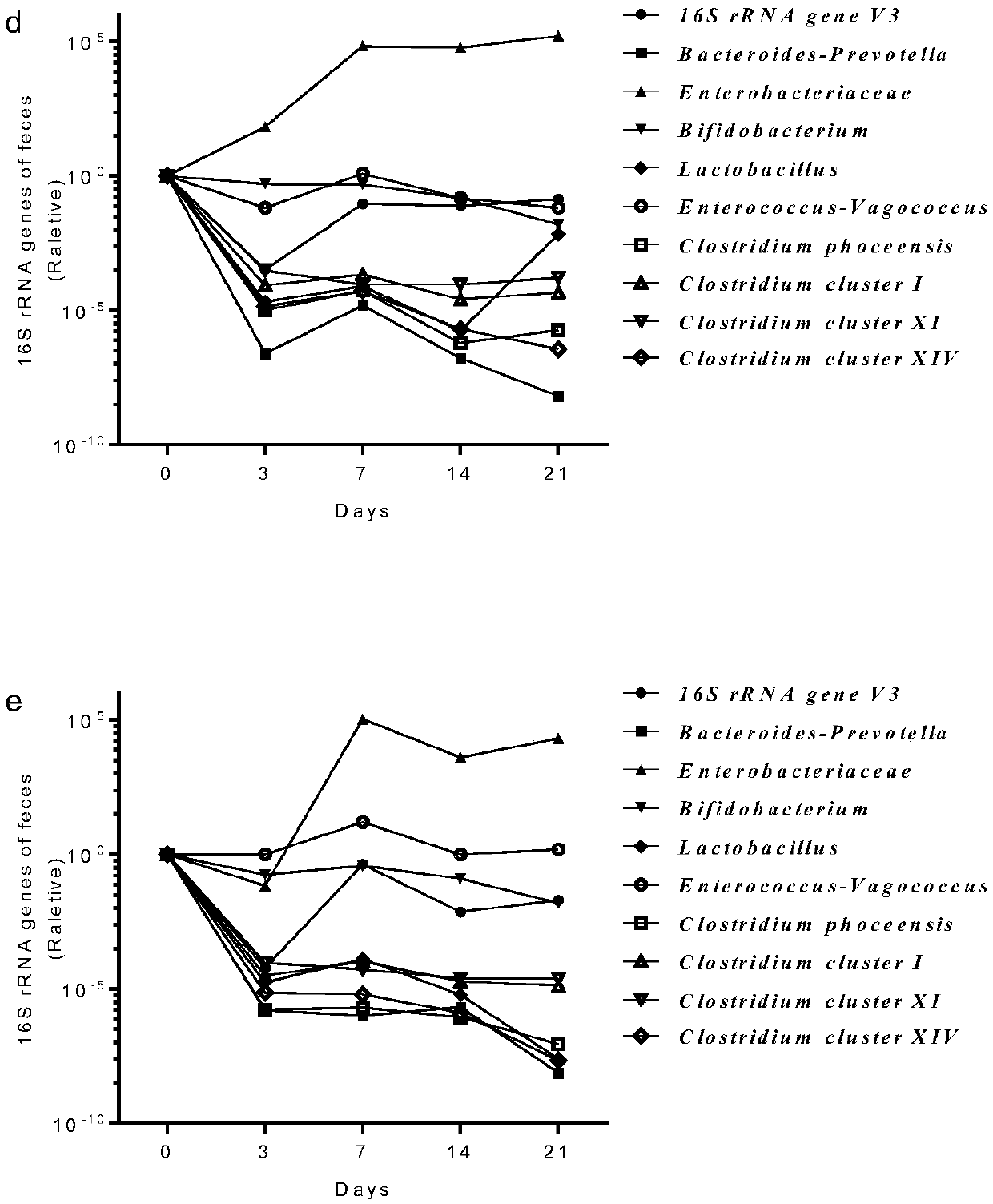
[0024] Establishment, evaluation and analysis of a non-rodent animal model for the study of gut commensal flora.
[0025] 1. Construct a primate model in which the intestinal commensal flora has been cleared
[0026] 1.1 Take 12-month-old rhesus monkeys as experimental animals, 2 males (numbered 1 and 2 respectively), with a body weight of 3kg±0.5kg. Raised in a barrier environment, fed the same diet. The daily food needs are sterilized by autoclaving and consumed within 72 hours; the drinking water is sterilized by autoclaving and consumed within 24 hours.
[0027] 1.2 Preparation of mixed antibiotics: the antibiotics used include aminoglycosides, penicillins, glycopeptides and nitroimidazole antibiotics, including neomycin, streptomycin, ampicillin sodium, kanamycin, metronidazole and Vancomycin, both in powder form. The dosages are: neomycin 60mg / kg / day, streptomycin 60mg / kg / day, ampicillin sodium 60mg / kg / day, kanamycin 50mg / kg / day, metronidazole 60mg / kg / day , Vancomyci...
Embodiment 2
[0069] Repeat Example 1, only the number of rhesus monkeys and the antibiotic formula in step 1.2 are different:
[0070] The numbers of the two rhesus monkeys in this embodiment are 3 and 4 respectively.
[0071] 1.2 Preparation of mixed antibiotics: the antibiotics used include aminoglycosides, penicillins, glycopeptides and nitroimidazole antibiotics, including neomycin, streptomycin, ampicillin sodium, kanamycin, metronidazole and Vancomycin, both in powder form. The dosages are: neomycin 60mg / kg / day, streptomycin 60mg / kg / day, ampicillin sodium 60mg / kg / day, kanamycin 50mg / kg / day, metronidazole 60mg / kg / day , Vancomycin 60mg / kg / day. The average dose was divided into 3 times.
[0072] 1.2.1 Antibiotic formula 1: Weigh the corresponding weights of neomycin, streptomycin, ampicillin sodium and kanamycin respectively and dissolve them in 5% sterile sucrose water, mix them well and put them in the medicine tube ; Another corresponding weight of vancomycin was dissolved in 5% ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com