Amplicon next-generation sequencing based small fragment insertion and deletion detection method and device
An insertion-deletion and next-generation sequencing technology, applied in the field of biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0022] According to a typical embodiment of the present invention, the device for obtaining mismatched bases includes: TMAP alignment software, which is used to compare the sequence of the target region with the reference genome, and form mismatches with bases that are not aligned with the reference genome. Matching; samtools software, used to establish the index of comparison files.
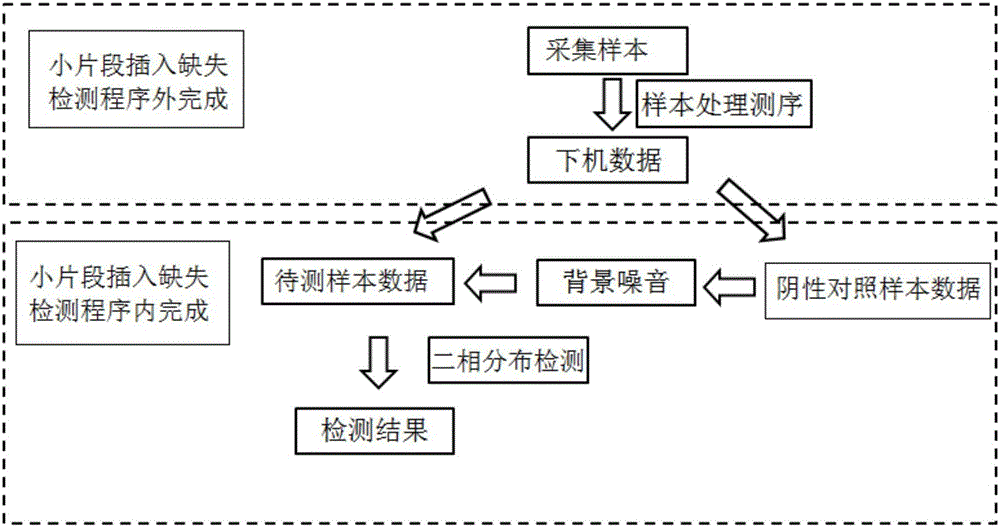
[0023] According to a typical embodiment of the present invention, the method for small fragment indel detection based on amplicon next-generation sequencing is as follows: figure 1 As shown, it mainly includes two parts completed outside the small fragment indel detection program and completed within the small fragment indel detection program. Among them, the former part includes sample collection, sample processing, sequencing and off-machine data; the latter includes negative control sample data processing The background noise is obtained, and the sample data to be tested and the background n...
Embodiment 1
[0027]In this example, the sample to be tested is the NCI-H1650 cell line (purchased from the American Type Culture Collection), which is a sample of a cell line that has a 0.21% frequency of EGFR:p.E746_A750delELREA positive mutation tested by 3d-PCR.
[0028] The specific steps of this embodiment are as follows (all reagents are purchased from Thermo Fisher):
[0029] 1) Extract DNA from the sample to be tested, quantify it with a fluorometer (Qubit), concentrate or dilute with nuclease-free water to make the concentration 5ng / ul and the volume 6ul.
[0030] 2) Multiplex PCR technology was used to amplify the target region, and the PCR reaction system is shown in Table 1.
[0031] Table 1
[0032] Reagent volume High-fidelity multiplex amplification reaction mix 4μl primer mix 10μl plasma cell-free DNA 6μl
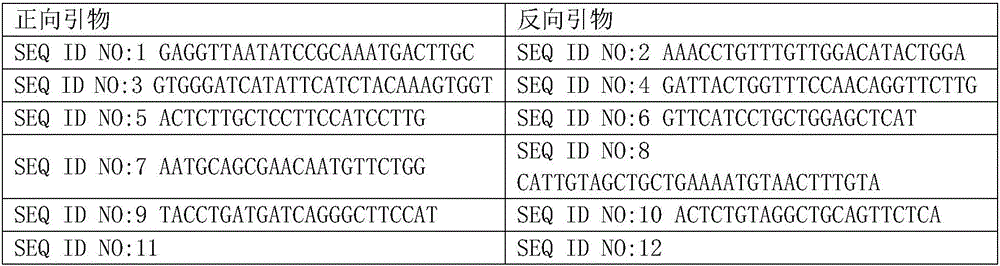
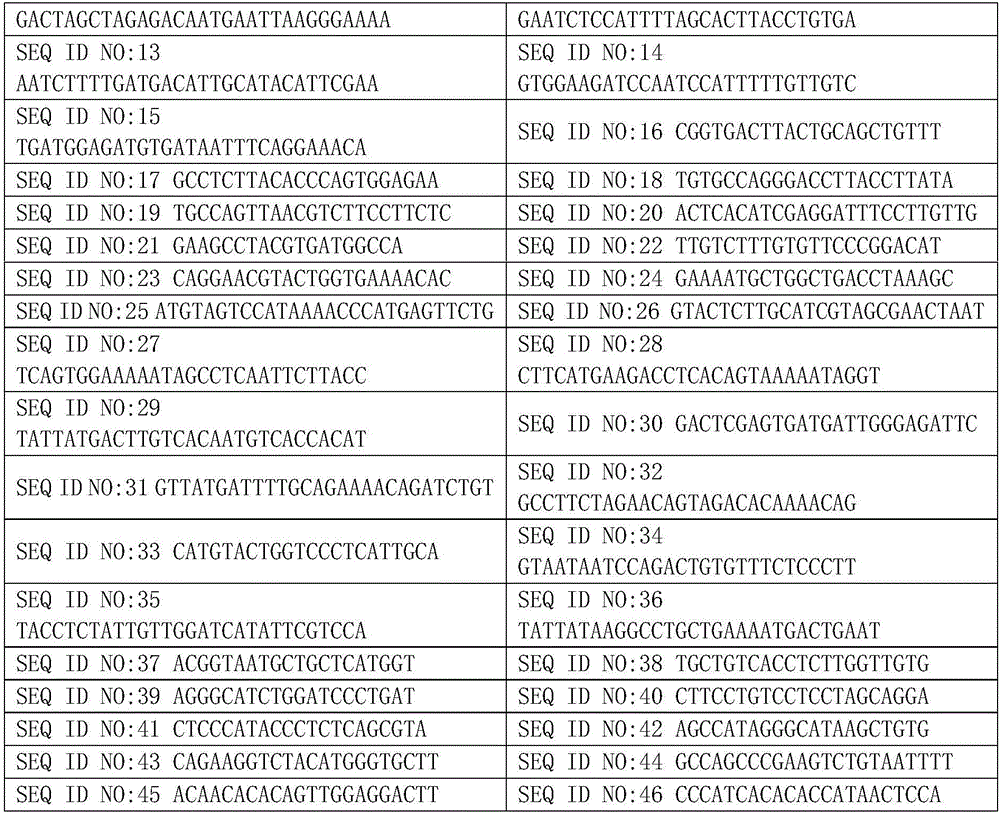
[0033] Among them, the specific sequences of the primers are shown in Table 2.
[0034] Table 2
[0035]
[0036]
[0037]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com