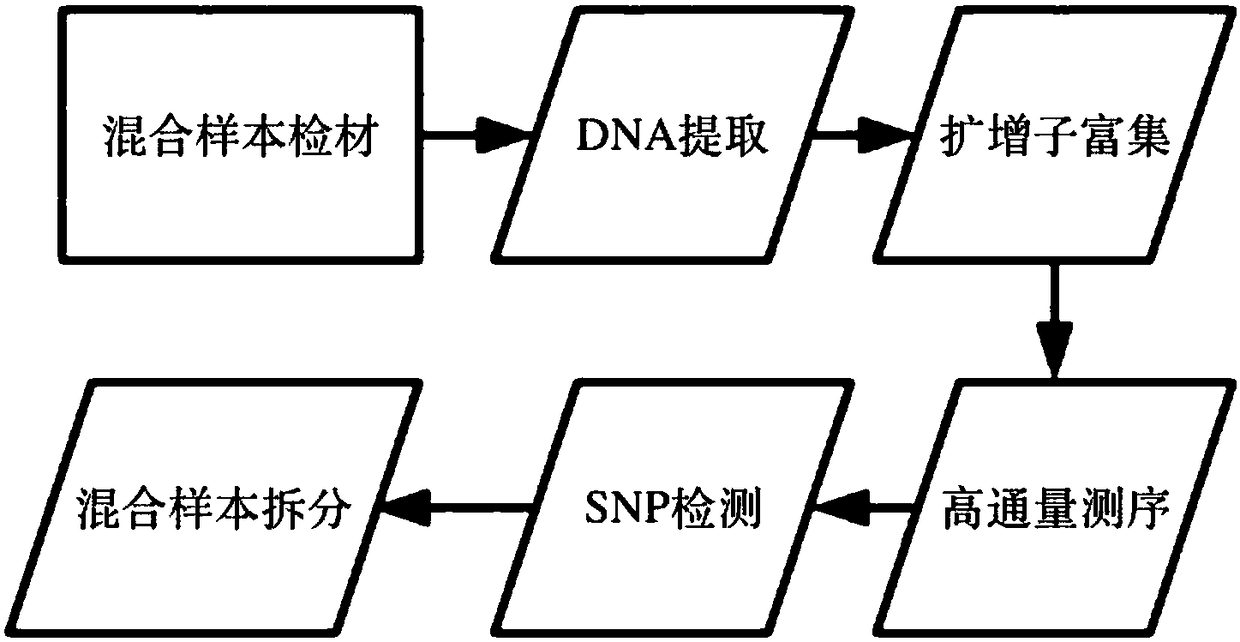
Method for separating mixed samples by SNP detection technology
A technology of mixed samples and detection technology, which is applied in the field of mixed sample splitting to achieve the effect of high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
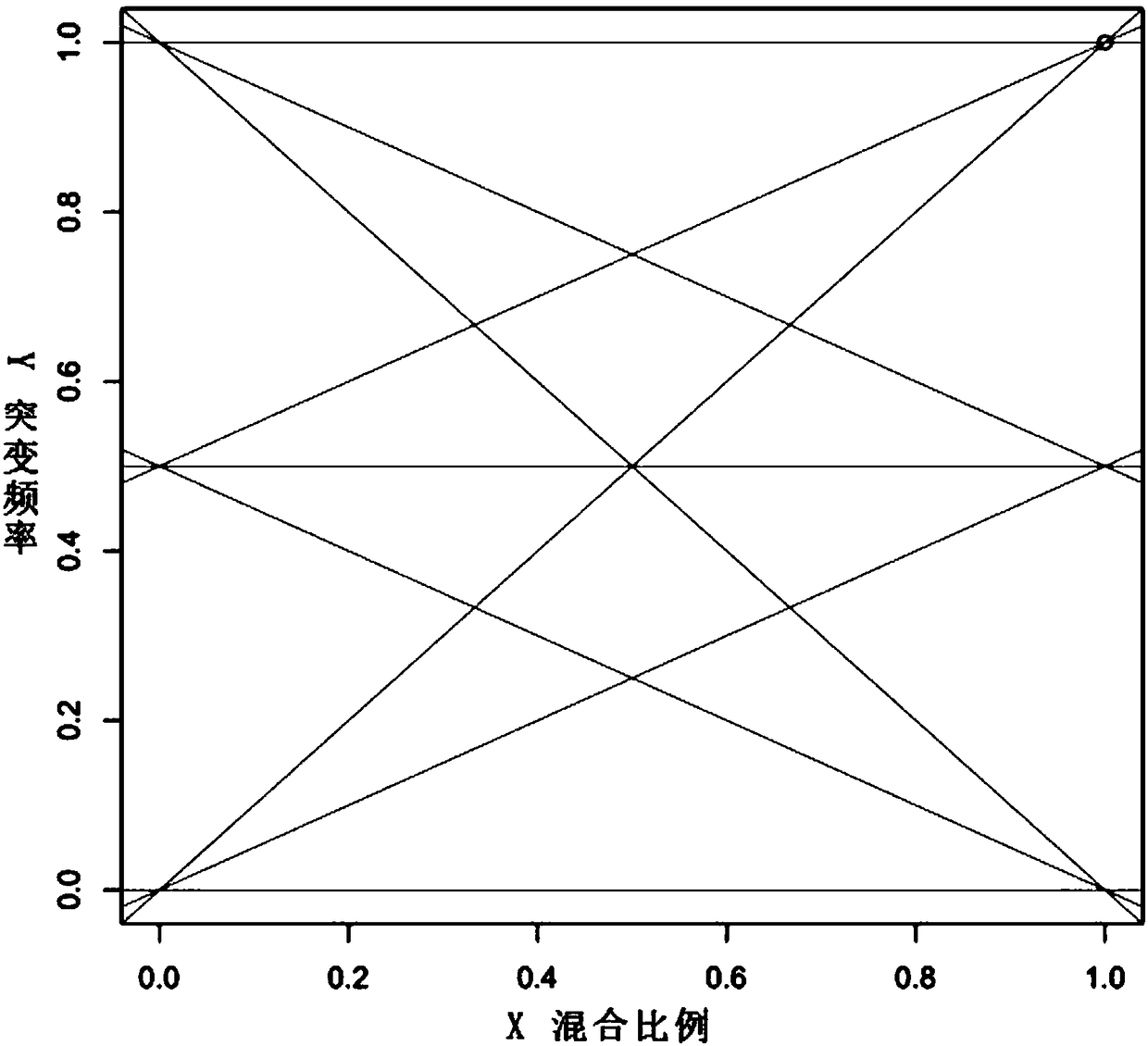
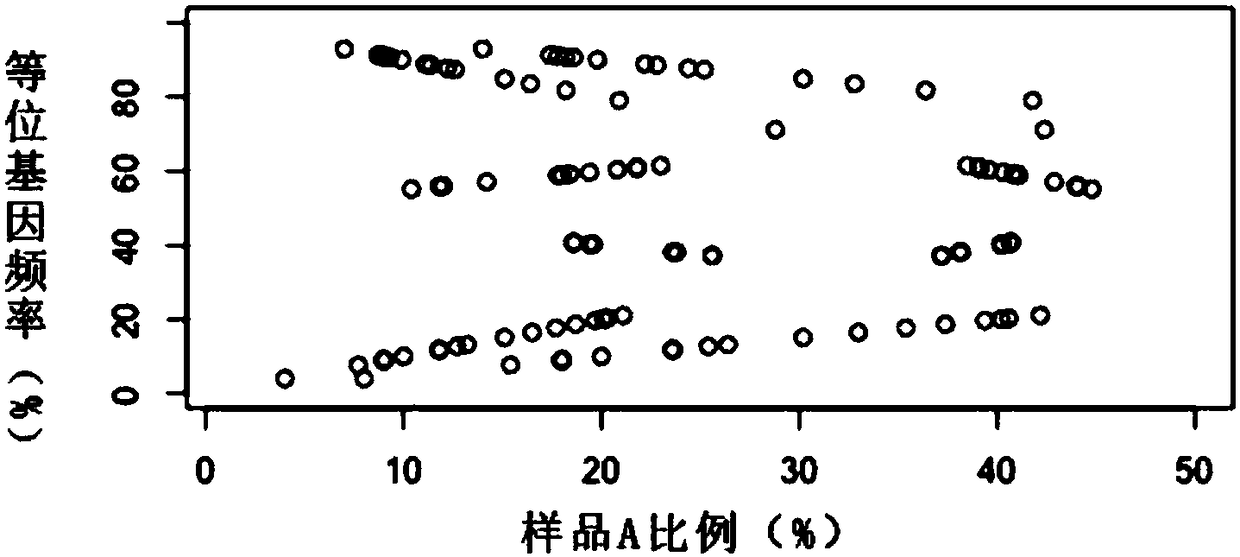
[0064] Example 1: Mixed sample ratio split
[0065] 1): Assume that the mixed sample includes two samples a and b, and the mixing ratio is X 0 :(100-X 0 ), X 0 0 >=0;
[0066] 2): Take N SNP sites to detect the mixed sample to obtain the SNP set of the mixed sample, obtain a total of 2N genotypes from the two samples a and b, and obtain the mutation frequency Y of each SNP site;
[0067] 3): For the mutation frequency Y of each SNP site, there are 9 mixing ratios of a and b, as follows:
[0068] a.2*Y=X 1 *GT 0 +(100-X 1 )*GT 0
[0069] b.2*Y=X 2 *GT 0 +(100-X 2 )*GT 1
[0070] c.2*Y=X 3 *GT 0 +(100-X 3 )*GT 2
[0071] d.2*Y=X 4 *GT 1 +(100-X 4 )*GT 0
[0072] e.2*Y=X 5 *GT 1 +(100-X 5 )*GT 1
[0073] f.2*Y=X 6 *GT 1 +(100-X 6 )*GT 2
[0074] g.2*Y=X 7 *GT 2 +(100-X 7 )*GT 0
[0075] h.2*Y=X 8 *GT 2 +(100-X 8 )*GT 1
[0076] i.2*Y=X 9 *GT 2 +(100-X 9 )*GT 2
[0077] Among them, GT 0 Represents no mutation as 0, GT 1 Represents ...
Embodiment 2
[0080] Embodiment 2: Public Security Criminal Evidence Appraisal
[0081] 1. DNA extraction: gDNA extraction of mixed samples of criminal cases.
[0082] 1) Take the mixed sperm spot (or the mixed blood spot on the murder weapon) in the case of sexual assault into a centrifuge tube containing cell lysate. Invert the centrifuge tube 5-6 times to mix.
[0083] 2) Incubate at room temperature for 10 minutes to lyse the cells. Centrifuge at 2000xg for 10 minutes at room temperature.
[0084] 3) Remove the supernatant, leaving about 1.4mL of liquid in the centrifuge tube.
[0085] 4) Add RNase, protein precipitation solution, and centrifuge at 2000xg for 10 minutes.
[0086] 5) Add isopropanol to the supernatant, centrifuge at 2000×g for 1 minute at room temperature, and discard the supernatant.
[0087] 6) Wash with ethanol and dry to obtain DNA.
[0088] 2. Library construction, template preparation
[0089] 1) Configure the DNA targeted amplification reaction system
[0...
Embodiment 3
[0102] Example 3: Non-invasive Fetal Paternity Test
[0103] In the existing forensic physical evidence identification, the identification of parent-child relationship requires the "child", "father", and "mother" to provide their own test materials, such as blood, hair, saliva, oral cells and bones, etc. can be used for parent-child test. materials, the "children" of which need to be independent individuals to be accurately sampled. The technology of this solution can isolate the SNP set of the child from the peripheral blood of the pregnant woman, eliminate the factor that the child is an independent individual, and determine the parent-child relationship as soon as possible. Since the concentration of fetal free DNA (cell-free-fetusDNA) in the peripheral blood free DNA (cell-free DNA) of pregnant women over six weeks is 5% to 15%, the peripheral blood of pregnant women can be regarded as the sample material mixed by two people. The method of splitting mixed samples by SNP d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com