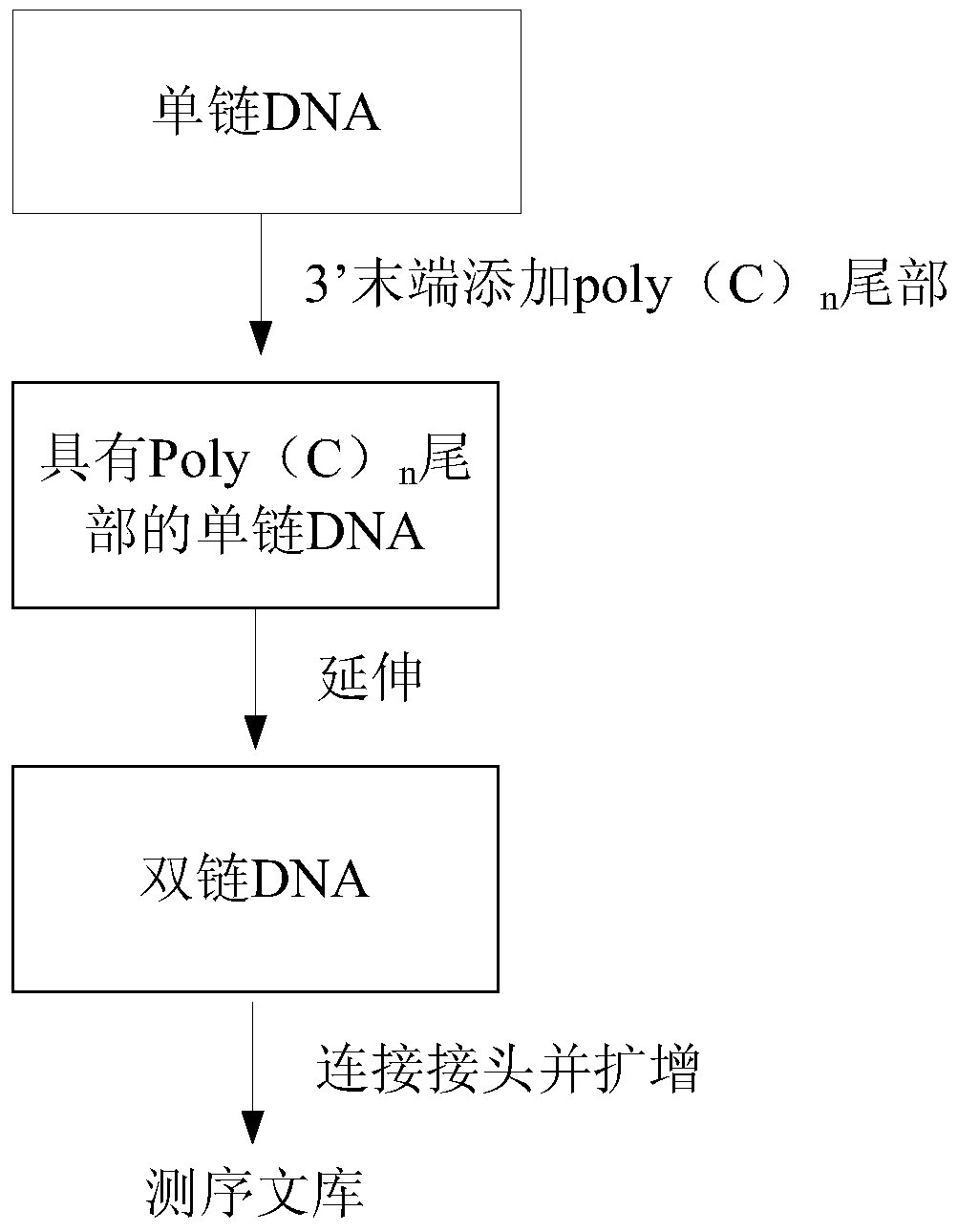
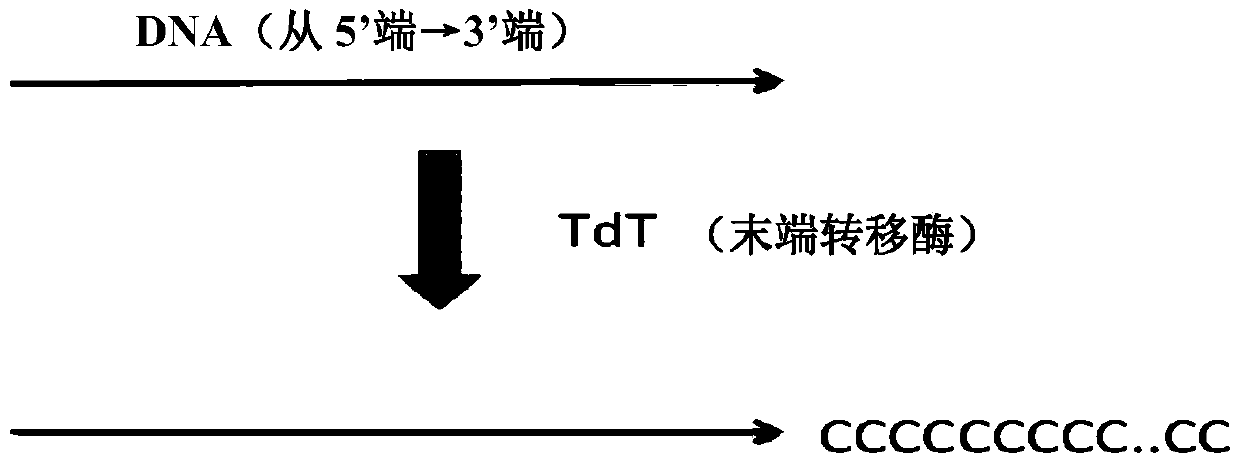
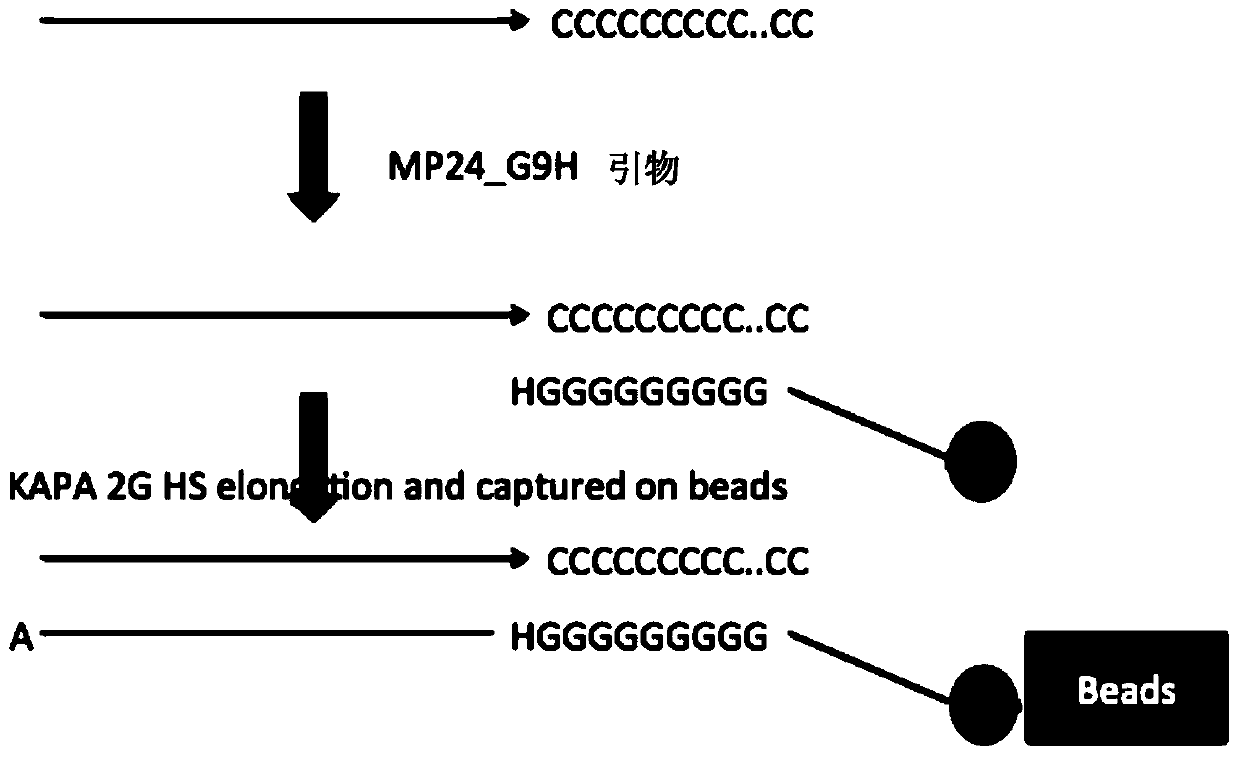
Method for constructing sequencing library based on single-stranded dna molecule and its application
A DNA molecule, sequencing library technology, applied in DNA preparation, recombinant DNA technology, library and other directions, can solve problems to be improved, and achieve the effect of keeping genetic information intact, efficient determination, and less sample loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0099] Example 1 Construction of a sequencing library
[0100] 1. The DNA used for library construction was accurately quantified by Qubit fluorometer (Invitrogen, Q32857). For double-stranded DNA, use Qubit dsDNA HS assay kit (Invitrogen, Q32854) for quantification; for single-stranded DNA, use QubitssDNA HS assay kit (Invitrogen, Q10212) for quantification. 25pg--10ng of DNA can be used as the initial amount for library construction, and then follow the TELP standard library construction steps described below.
[0101] For a small amount of double-stranded DNA (such as ChIP-Seq DNA) that is mechanically broken (such as ultrasonically broken), first perform end repair on the 5' and 3' ends of the mechanically damaged DNA. The reaction system is: 32.6 μl DNA sample , 4 μl 10x T4ligase buffer (NEB, B0202S), 1.6 μl 10 mM dNTP mix (NEB, N0447S), 0.8 μl T4PNK (NEB, M0201S), 0.8 μl T4 DNA polymerase (NEB, M0203S) and 0.16 μl Klenow fragment (NEB, M0210S) Mix well and react at 20°...
Embodiment 2
[0127] Example 2 RNA-Seq (cDNA sequencing)
[0128] Implement the present embodiment according to the steps identical with embodiment 1, only difference is:
[0129]Total cellular RNA was extracted with TRIzol reagent (Invitrogen), purified with microPoly(A) Purist Kit (Ambion, AM1919), and excess DNA was digested with DNase I, and poly-T primer (T18) and M-MLVReverse Transcriptase (Invitrogen) were used to Perform reverse transcription. After being broken to a suitable length by ultrasound, RNaseA is added to digest the RNA template strand, leaving the cDNA strand, and the single-stranded cDNA is obtained after purification. At this point, the single-stranded cDNA can be standardized using TELP technology to build a library (single-stranded DNA can skip the step of end repair and directly perform Tailing and the following steps), thereby retaining the specificity of the strand.
Embodiment 3
[0130] Example 3ChIP-Seq (chromatin immunoprecipitation sequencing)
[0131] Implement the present embodiment according to the steps identical with embodiment 1, only difference is:
[0132] Cells used for ChIP were first cross-linked with 1% formaldehyde at 37°C for 10 min, and the cells were lysed to release the chromatin. Fragments of 200-500 bp were fragmented by ultrasound. For each ChIP reaction, add 2-5 μg of the corresponding antibody and incubate overnight at 4°C. Finally, the enriched double-stranded DNA fragments were obtained, quantified by Qubit, and a certain amount of DNA was taken for TELP standardization library construction. For the application of ChIP-Seq, the library construction steps are exactly the same as those described in Example 1. Our experiment proves that this method can be used to successfully and efficiently build a standard library for DNA as low as 25pg-1ng.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com