DU5 adaptor, application thereof and cDNA library constructed by dU5 adaptor
An adapter and library technology, applied in the field of cDNA library, can solve the problems of long library construction time, large amount of RNA input, and low PCR efficiency, so as to reduce the formation of adapter-primer dimers, expand the scope of research, reduce The effect of cycle number
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] This embodiment provides a dU5' adaptor, which has the following nucleotide sequence: 5'- / 5Phos / -AGATCGGAAGAGCGTCGTGTAGC / dU / CTTCCGATCTNNNNNNNNNN- / 3SpC3 / -3' (SEQ ID NO: 3).
[0057] The dU5' adaptor is a structure containing deoxyuracil specially designed by the inventor. In the newly designed dU5' adaptor, the insertion position of dU is the 24th position from the 5' end, which is more conducive to subsequent PCR The forward primer is paired, and its complementary pairing part is kept intact, which is conducive to the binding of the forward primer and the template, can effectively reduce the formation of adapter-primer dimer, and reduce the number of cycles required for the reaction, thereby improving PCR amplification. Increase efficiency, thereby improving the quality of the library.
[0058] The dU5' adaptor in this example was obtained by commercial synthesis according to its nucleotide sequence, and the commercially synthesized dU5' adaptor nucleic acid sam...
Embodiment 2
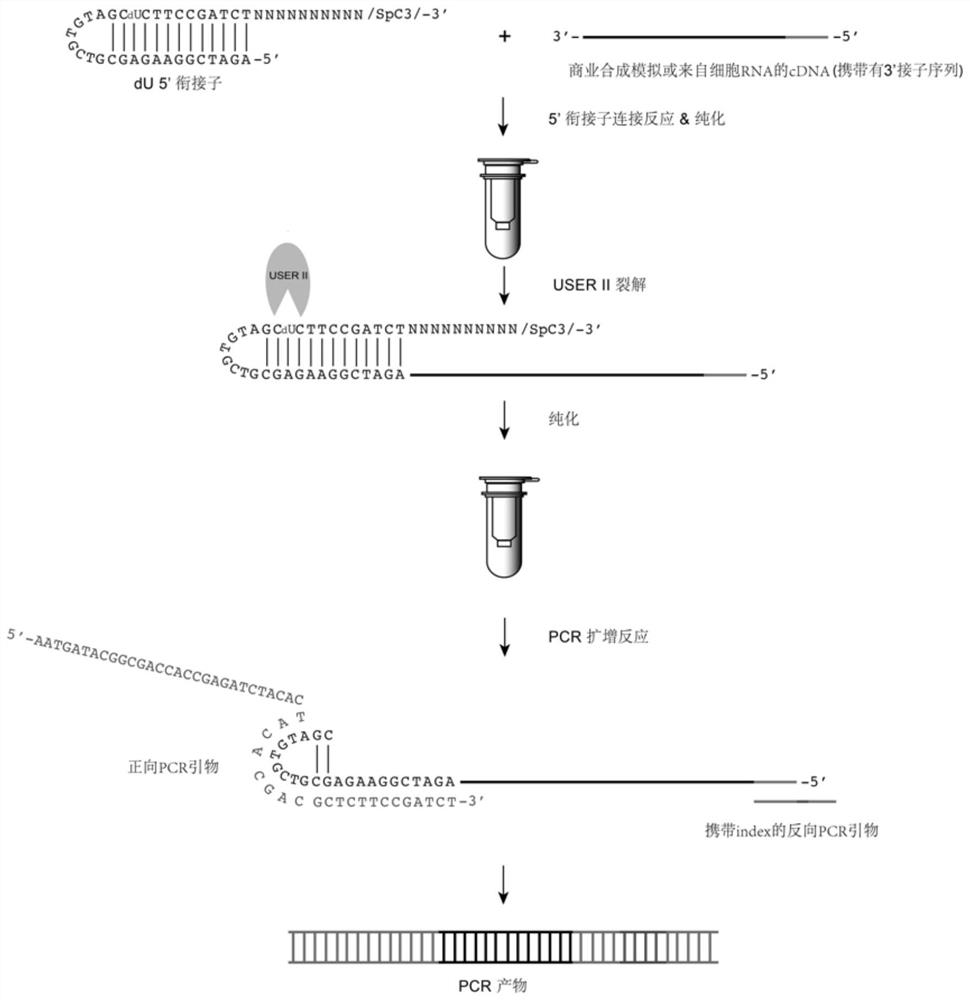
[0060] The present embodiment provides a method for constructing an improved cDNA library. The improved part is as follows: figure 1 shown, it includes the following steps:
[0061] The simulated cDNA nucleic acid sample (which has a 3' adaptor, see SEQ ID NO: 4 for the sequence) was mixed with the dU 5' adaptor of Example 1 to carry out a ligation reaction using a rapid ligation kit (NEB, M2200L). The molar ratio of the nucleic acid sample to the dU5' adaptor was 1:10, and the reaction conditions were 37°C for 2 h; the dU5' adaptor-cDNA nucleic acid complex was obtained and subjected to column purification (Zymo Research, R1016);
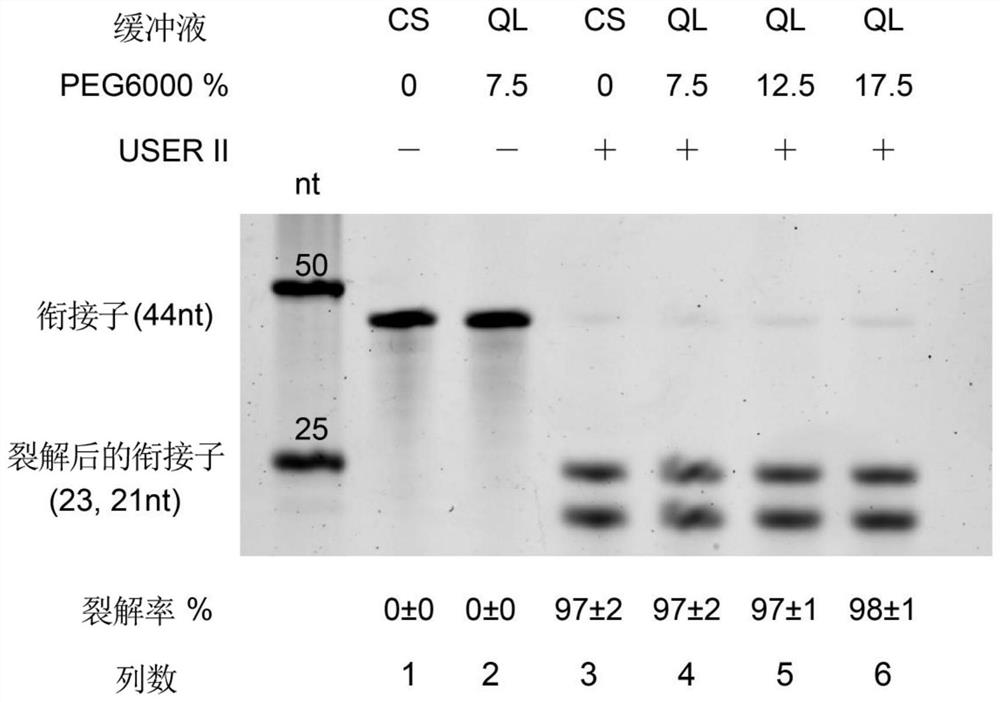
[0062] Add CutSmart buffer (1X final) USERII enzyme (0.1U / μL final) to the purified dU5' adaptor-cDNA nucleic acid complex to cleave deoxyuracil at a cleavage temperature of 37°C and a reaction time of 15min to form ends Partially complementary adaptor-nucleic acid complexes; excess adaptors were removed by column purification (Zymo Research, R10...
Embodiment 3
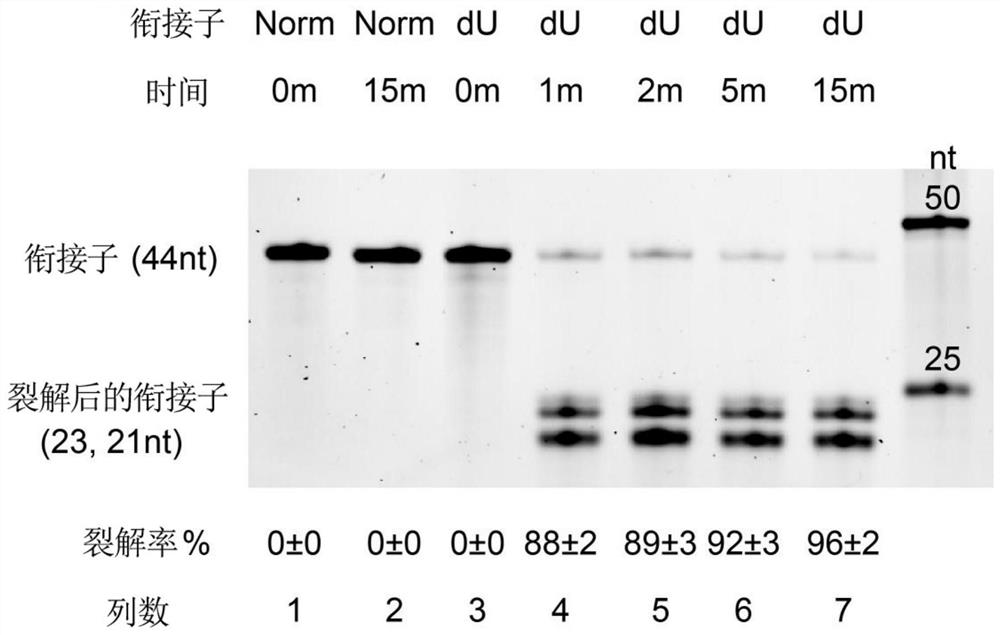
[0072] Example 3: Cleavage time optimization of deoxyuracil in dU 5' adaptors by USER II enzyme
[0073] This example investigates the effect of USER II enzyme on the cleavage of deoxyuracil in the cleavage of dU5' adaptors at different cleavage times, and the reaction conditions are set to 37 °C. The experimental results are as figure 2 shown.
[0074] Depend on figure 2 It can be seen that the cracking rate increases with the increase of the reaction time. In order to maximize the cracking rate, a cracking time of 15min is preferably used.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com