A kit and method for detecting chromosome heterozygosity loss based on amplicon sequencing
A loss of heterozygosity, kit technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of chromosome deletion, non-disclosure and implication, high and low gene expression, etc. Achieve the effect of increasing the amount of DNA and reducing the cost of detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1 kit of the present invention
[0026] The kit for detection of chromosomal heterozygosity loss based on amplicon sequencing of the present invention includes:
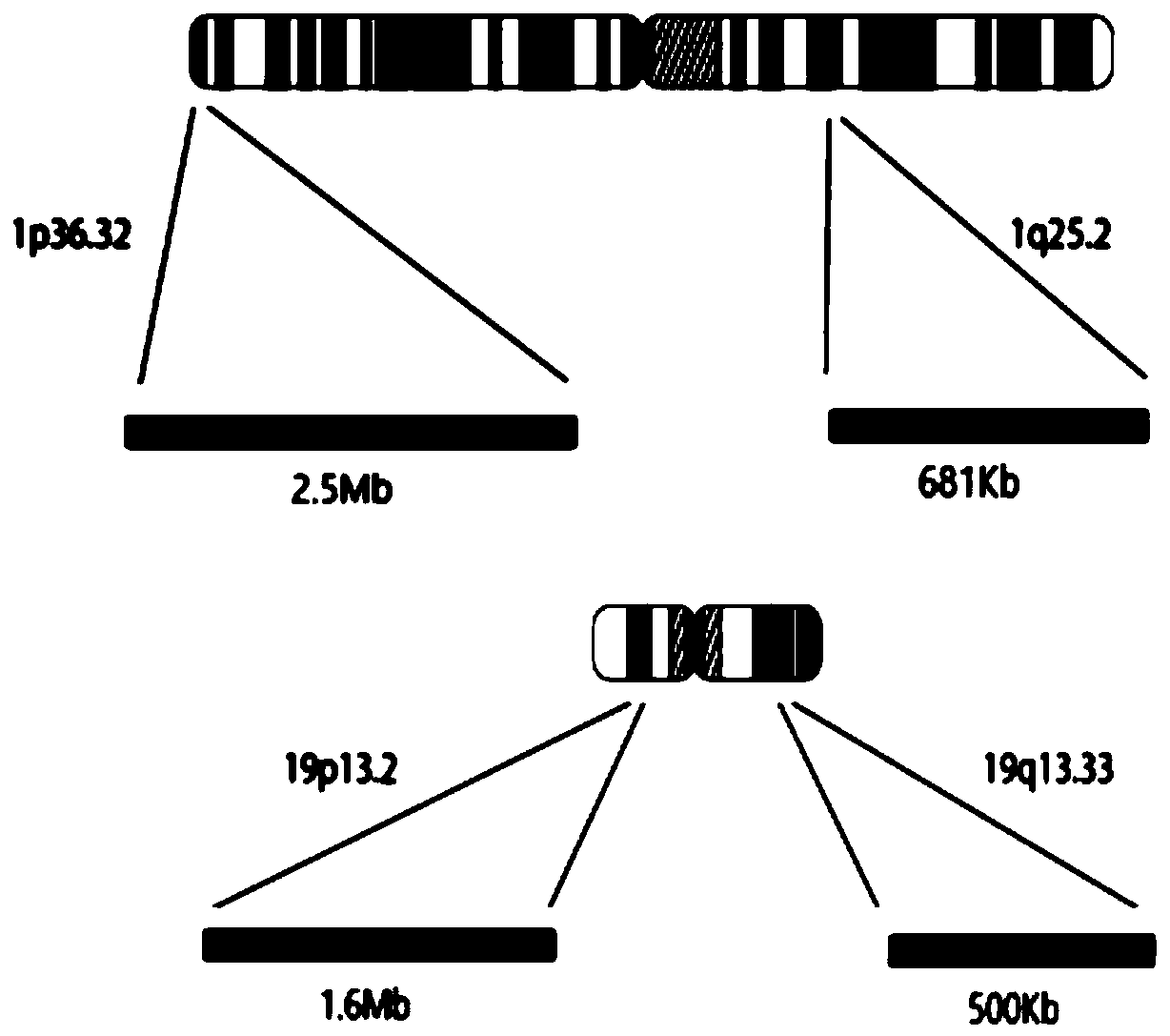
[0027] A primer pair set for determining the expression of 14 genes in the 1p36.32 region of the short arm of human chromosome 1: the primer pair set consists of SEQ ID NO: 1 and SEQ ID NO: 22, SEQ ID NO: 2 and SEQ ID NO: 23, SEQ ID NO: 3 and SEQ ID NO: 24, SEQ ID NO: 4 and SEQ ID NO: 25, SEQ ID NO: 5 and SEQ ID NO: 26, SEQ ID NO: 6 and SEQ ID NO: 27, SEQ ID NO : 7 and SEQ ID NO: 28, SEQ ID NO: 8 and SEQ ID NO: 29, SEQ ID NO: 9 and SEQ ID NO: 30, SEQ ID NO: 10 and SEQ ID NO: 31, SEQ ID NO: 11 and The primer composition of SEQ ID NO: 32, SEQ ID NO: 12 and SEQ ID NO: 33, SEQ ID NO: 13 and SEQ ID NO: 34, SEQ ID NO: 14 and the sequence shown in SEQ ID NO: 35, primer sequence see Table 1 The primer sequences of 14 genes in the 1p36.32 region of the short arm of human chromosome 1, and the concentration...
Embodiment 2
[0049] Example 2 Detection of 1p19q loss of heterozygosity in paraffin-embedded section samples from patients with glioma
[0050] 1. Detection method
[0051] 1.1 Extract DNA from paraffin sections.
[0052] 1.2 The components of the first PCR amplification (multiple targeted PCR amplification) are as follows:
[0053] components Final concentration primer mix 0.2μM / each GC stabilizer 1× PCR Enzyme Mix 1× FFPE DNA template 10ng wxya 2 o
to 25μL
[0054] Primer mixture preparation: 88 kinds of 1p36.32, 1q25.2, 19p13.2, 19q13.33 regions described in Table 1, Table 2, Table 3, and Table 4 described in Example 1, the concentration is 100 μM Add 3.5 μL of each primer solution to a centrifuge tube and mix thoroughly. When in use, take 10 μL and add it to the above PCR reaction system. The final concentration of each primer is 0.2 μM.
[0055] 1.3 The first PCR reaction procedure:
[0056]
[0057] 1.4 After the program runs,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com