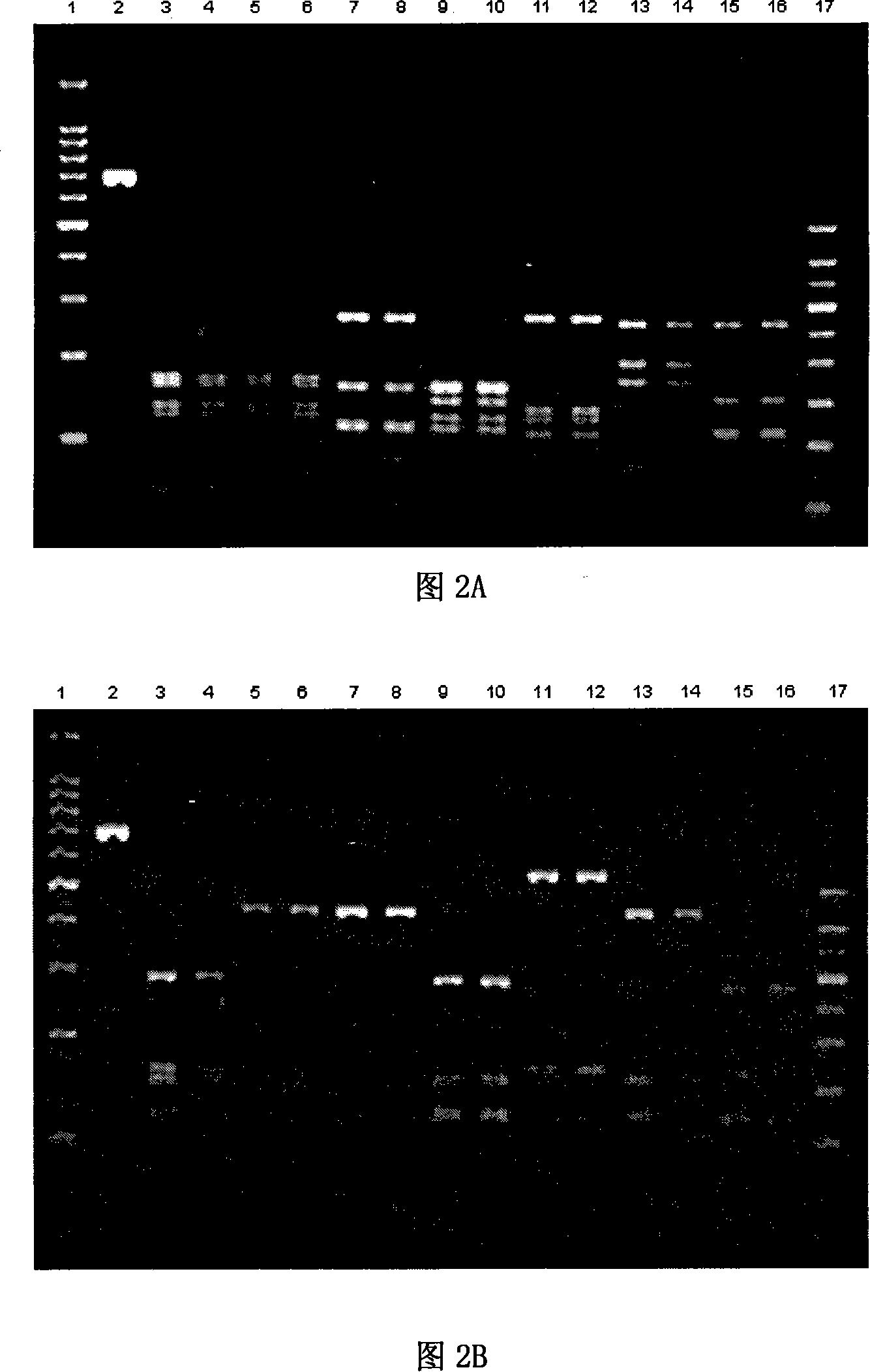
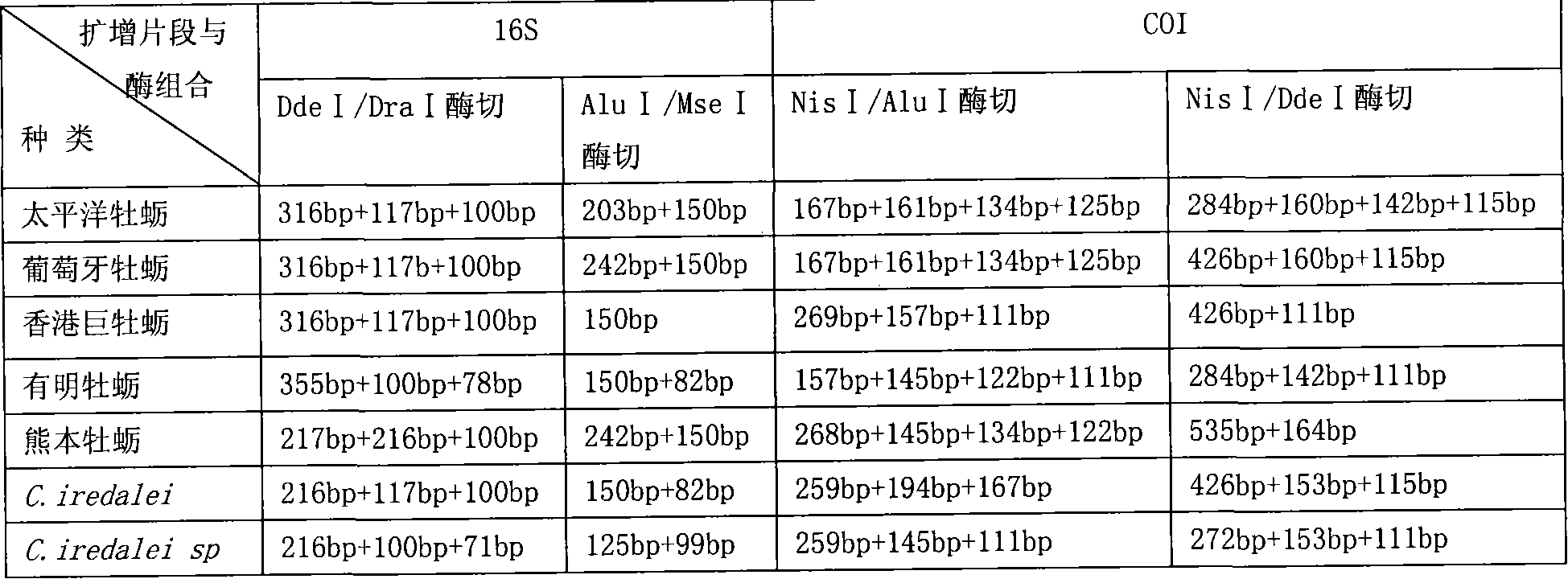
PCR-RFLP identification method for seven crassostrea oyster on south China coast
A technique of PCR-RFLP and giant oyster, applied in the field of molecular biology, achieves the effect of high reliability and wide application range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0052] Example 2 Using COI Fragment PCR-RFLP to Identify the Species of Giant Oyster Ariake
[0053] (1) Genomic DNA extraction
[0054] Take 0.1 g of adductor muscle tissue, add 700ul extraction buffer to digest overnight at 37°C; use an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) mixture, shake and mix well, 12000g / and centrifuge for 10 minutes; take the supernatant and repeat the previous step once; add an equal volume of chloroform, mix well, and centrifuge at 12000g / min for 5 minutes; take the supernatant and add 2 times the volume of absolute ethanol and 1 / 10 Sodium acetate with a volume of 3M; 12000g / min, centrifuge for 12 minutes, remove the supernatant, and wash with 70% ethanol for 1-2 times; after the ethanol is completely evaporated, dissolve the genomic DNA precipitate in 50ul sterile purified water to obtain genomic DNA. Store at 4°C for later use.
[0055] (2) PCR amplification of COI fragment
[0056] The PCR reaction system is 50ul, includi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com