Process for abstracting total DNA of swine waste sample
An extraction method and feces sample technology, applied in the field of microbiology and molecular biology, can solve the problems of complex composition of animal feces, difficulty in selecting glass filters, and difficulty in popularization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1: The method of the present invention extracts the total DNA of a swine feces sample.
[0036] The samples used in the experiment were the collected feces of Rongchang pigs, and the collected feces samples were stored at -20°C for future use. Specific steps are as follows:
[0037] (1) Weigh an appropriate amount of stool sample into a sterile 50ml centrifuge tube A, add 10-20ml PBS buffer solution and sterile glass beads and vortex to mix;
[0038] (2) After centrifuging at 200-400xg for 2-8 minutes, take out as much supernatant suspension as possible into another sterile 50ml centrifuge tube B;
[0039] (3) Wash the precipitate obtained in step (2) again with an appropriate amount of PBS buffer, centrifuge at 200-400xg for 2-8min, take out the upper layer suspension and combine it into centrifuge tube B;
[0040] (4) Add an appropriate volume of paraformaldehyde solution of a certain concentration to the centrifuge tube B, and incubate it at 4°C for 1-2 ...
Embodiment 2
[0053] Embodiment 2: The comparison between the present invention and the conventional method and kit method for extracting total DNA from feces.
[0054] In order to compare the parallel effect of the present invention with the existing conventional method and the commercial kit extraction method, the applicant carried out a comparison experiment using the method of the present invention, the conventional method and the kit method to extract at the same time, using the conventional method to extract total DNA from feces : According to the method reported by Rousselon et al. (see: Rousselon N, Delgenès JP, Godon JJ.J Microbiol Methods, 2004, 59(1): 15-22). Utilize kit method to extract total DNA from feces: the kit used is purchased from QiaGen company, and the commodity name of kit is DNA Stool Mini Kit. The feces samples used in the above two methods are the same as the present invention, and the extracted DNA is dissolved in 100 ml of TE buffer.
[0055] Adopt the method...
Embodiment 3
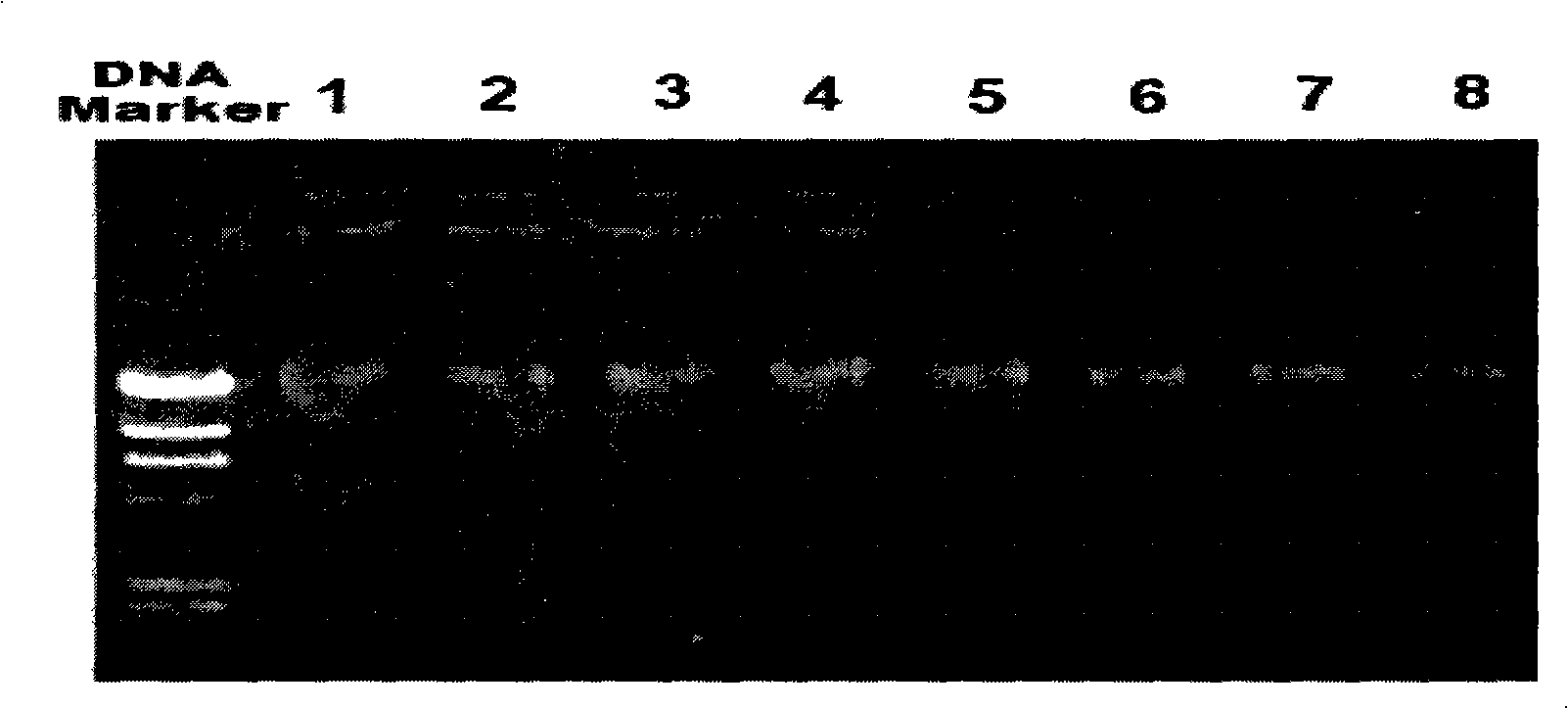
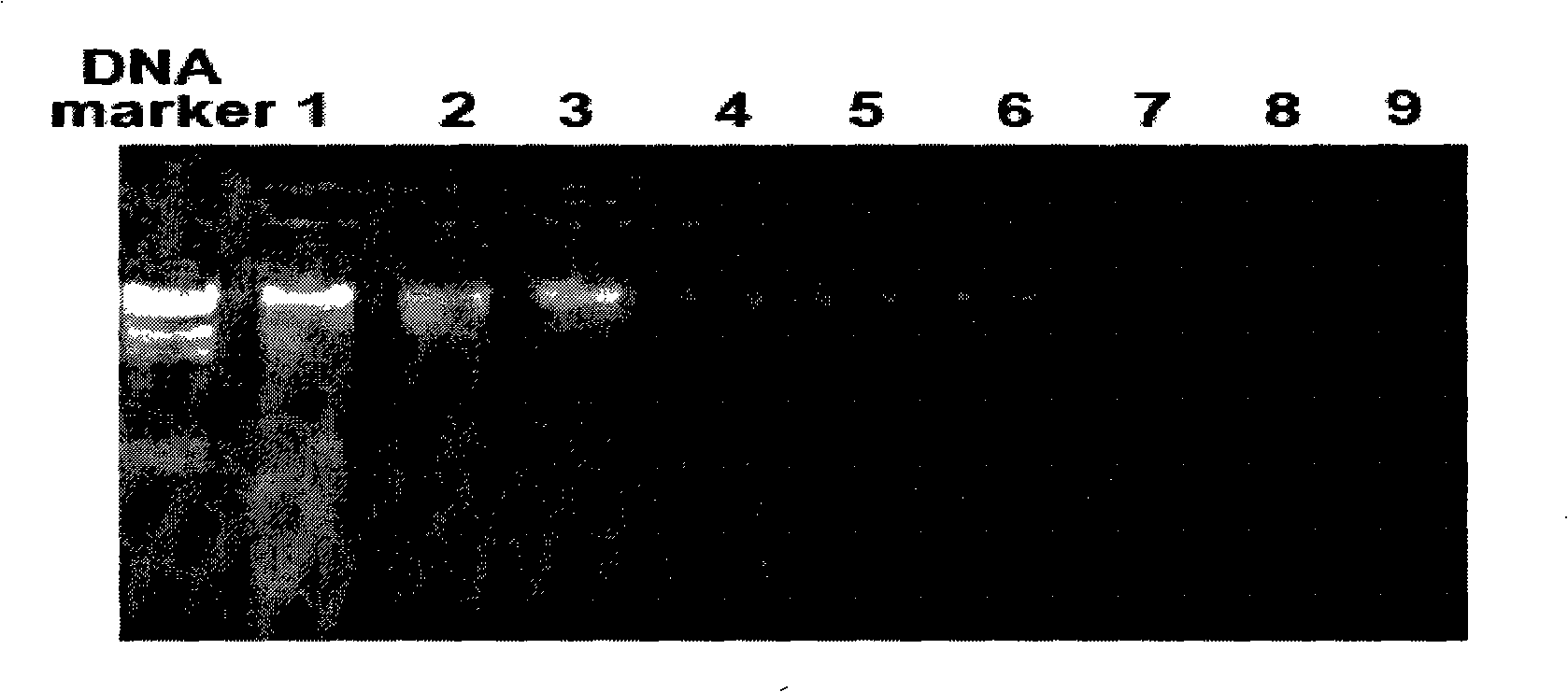
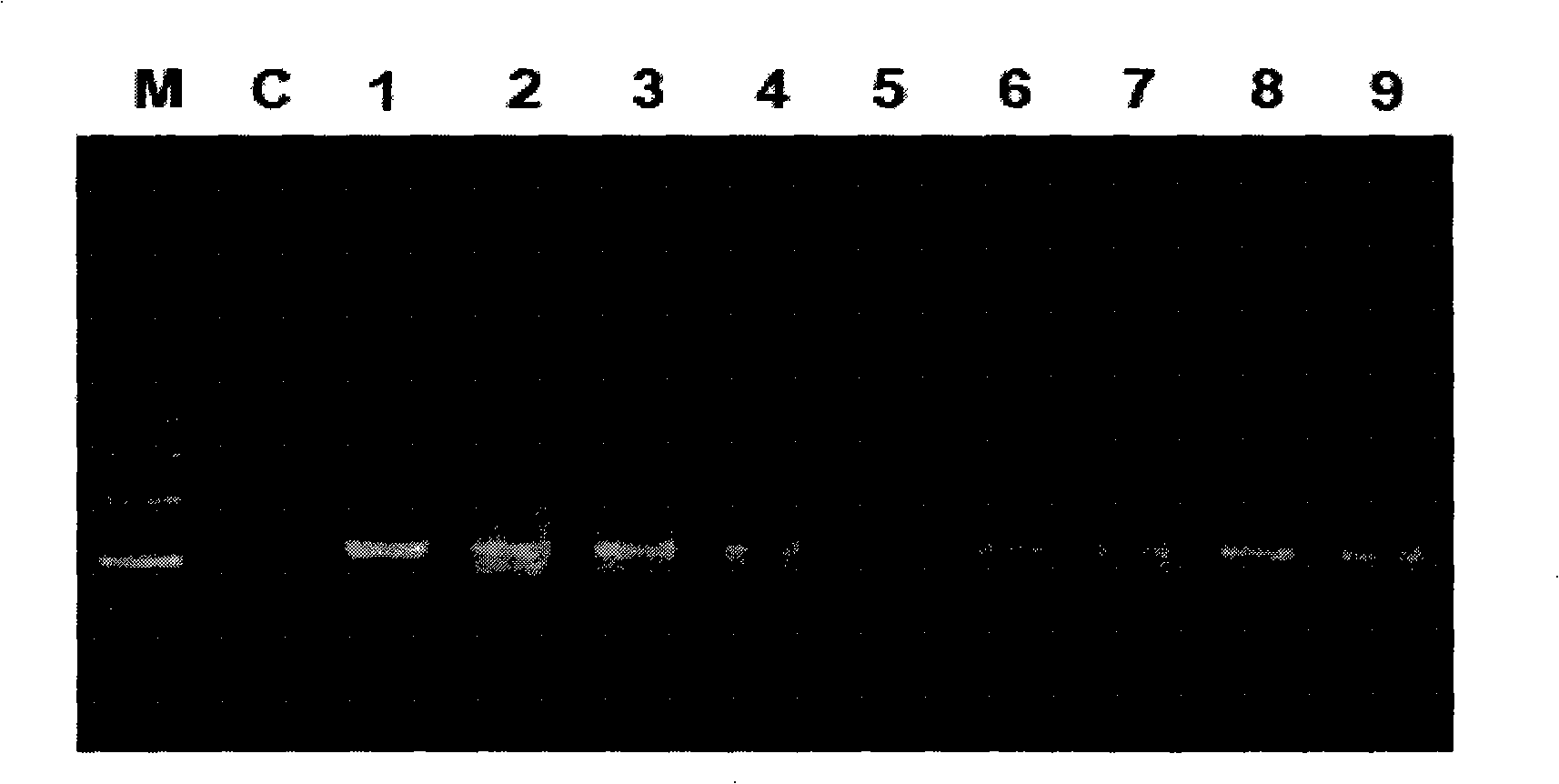
[0056] Example 3: The DNA extracted in Example 2 was subjected to PCR amplification detection of 16S rDNA.
[0057] Using the DNA extracted in Example 2 as a template, with the designed primers: the forward primer P0 is 5'-GAGAGTTTGATCCTGGCTCAG-3' and the reverse primer 1492r is 5'-CGGCTTACCTTGTTACGACTT-3' to amplify the 16S rDNA (16S ribosomal RNA gene). Utilize 1 μl of feces total DNA prepared by the conventional method, the kit method and the method of the present invention as a template for PCR reaction to amplify the 16S rDNA of fecal microorganisms respectively. The PCR reaction system is as follows (20 μl): 10×PCR buffer, 2 μl; 2.0 mMMgCl2; 200 μM dNTP; 0.5 μM primer; 1U Taq enzyme (TaKaRa); DNA template, 1 μl; add ddH20 to 20 μl; ℃ 5min; denaturation: 95℃ 30s; annealing: 58℃ 30s; extension: 72℃ 1.5min; final extension 72℃ 8min; 30 cycles.
[0058] After amplification, take 8 μl and perform electrophoresis detection on a 1% agarose gel. The size of the amplified targe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com