Three-level flow sequence comparison method based on many-core co-processor
A sequence comparison and processor technology, applied in the field of sequence comparison in the field of bioinformatics, can solve problems such as occupying program running time and affecting program running efficiency, so as to reduce comparison time, improve comparison efficiency, and increase comparison speed Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0055] The National Defense University uses a server equipped with two eight-core 2.4GHz CPUs and a 57-core 1.1GHz MIC card as the environment. The server hard disk size is 43TB, the memory size is 132GB, and the storage space on the MIC card is 6GB. The input data is the human genome. The space occupied by the reference genome is 3GB, and the space occupied by short DNA sequences is 240GB, including 80 million sequences, to verify the effect of the present invention:
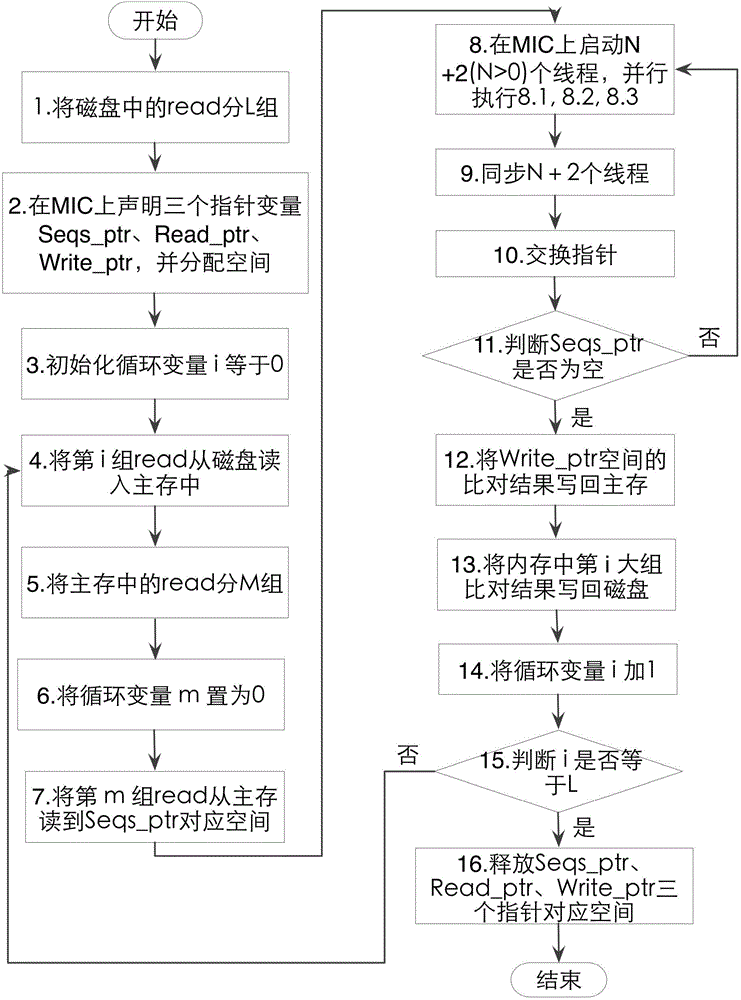
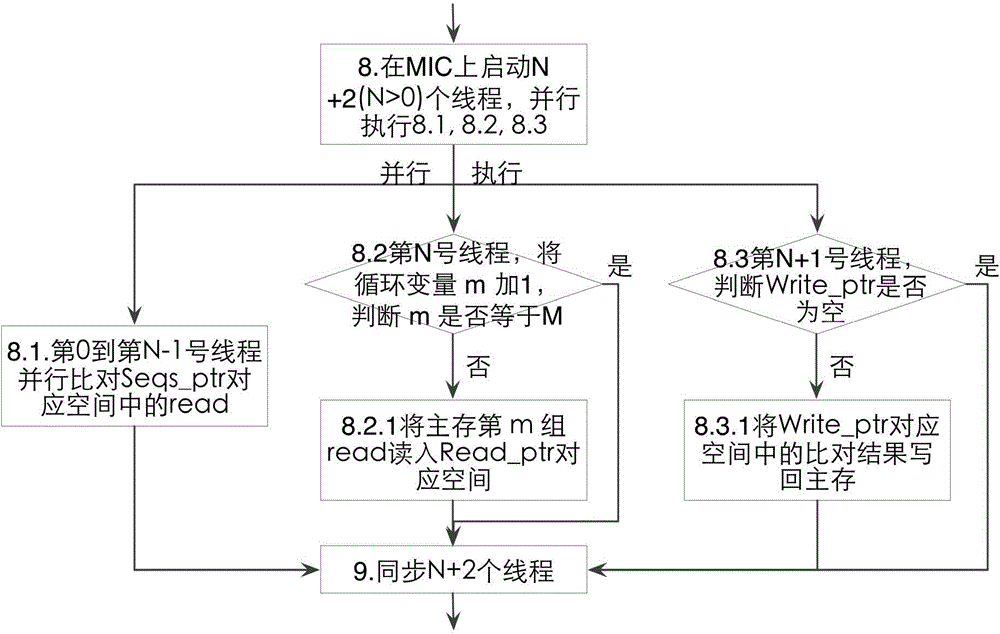
[0056] Such as figure 1 As shown, the specific implementation steps are as follows:
[0057] Step 1: The CPU is based on the available space of the main memory of the computer M_CPU=45GB (the operating system and other services occupy a certain amount of memory, and a part of the memory needs to be reserved for use when the program is running, so the available memory size is 45GB, which is less than the installed memory size of 132GB ), and the space occupied by the short DNA sequence M_DNA=240GB, the short DN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com