Next-generation sequencing short sequence rapid alignment analysis method and device
A second-generation sequencing and analysis method technology, applied in the field of bioinformatics, can solve the problems of low comparison efficiency and high memory usage of sequencing data, and achieve the effects of shortening comparison time, fast comparison speed and improving resource efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
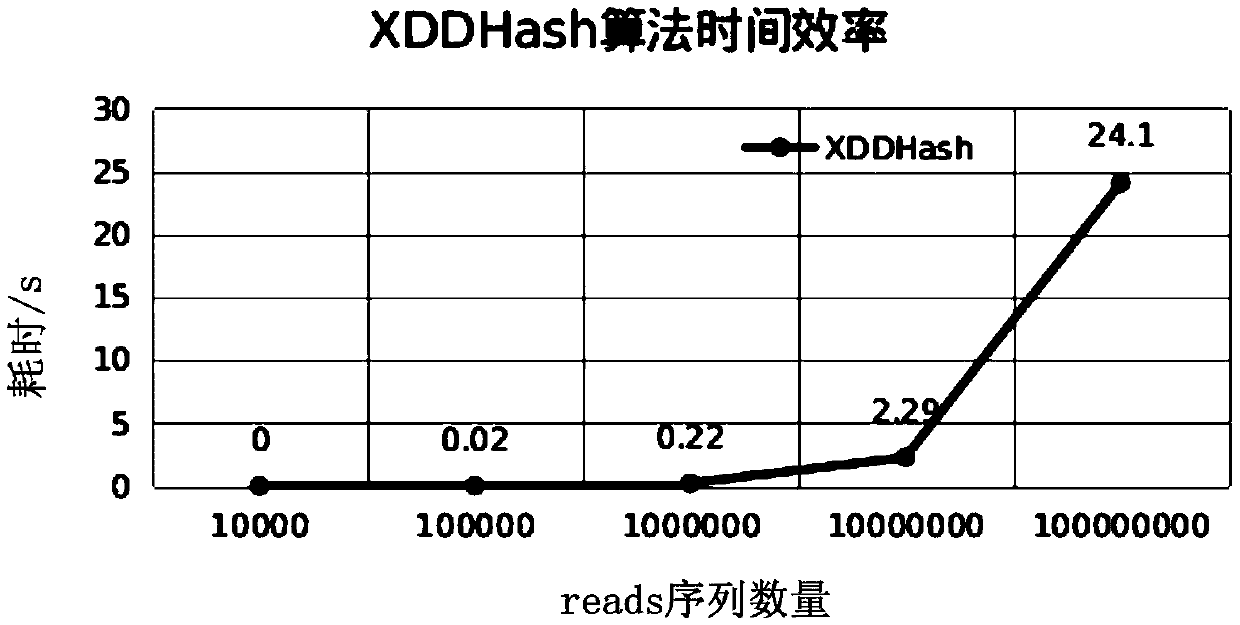
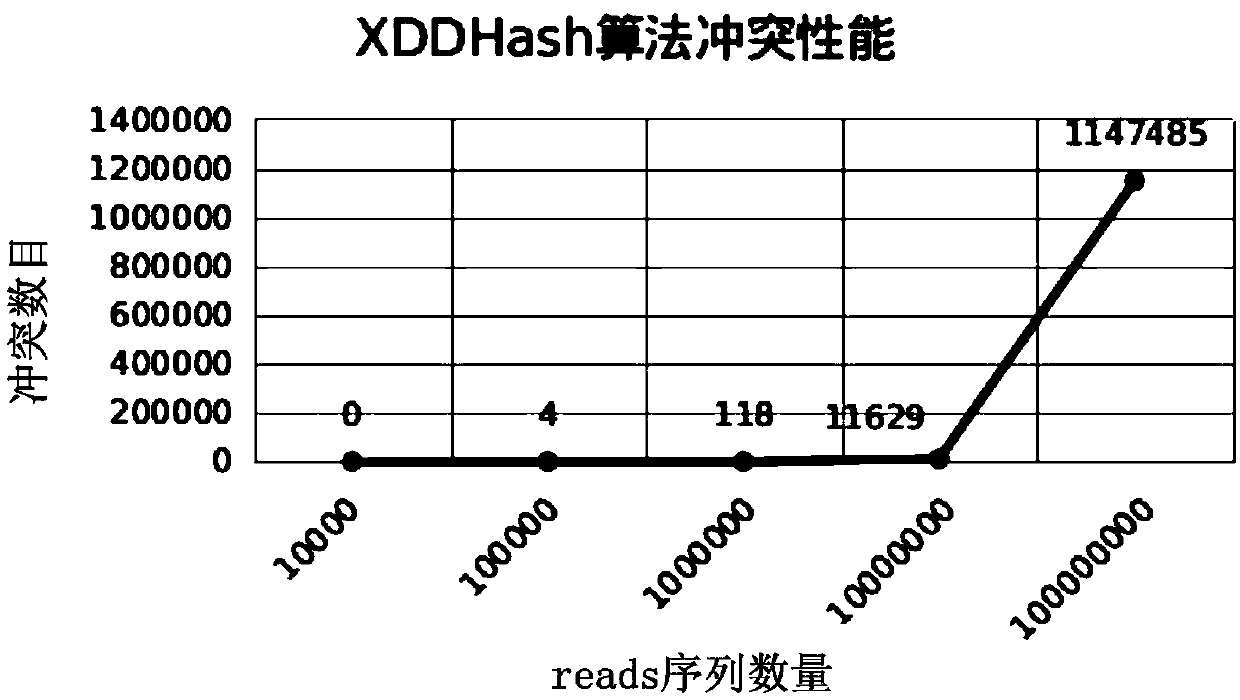
[0095] Example 1 Data analysis of non-invasive prenatal testing based on the rapid comparison and analysis method of short sequences of next-generation sequencing
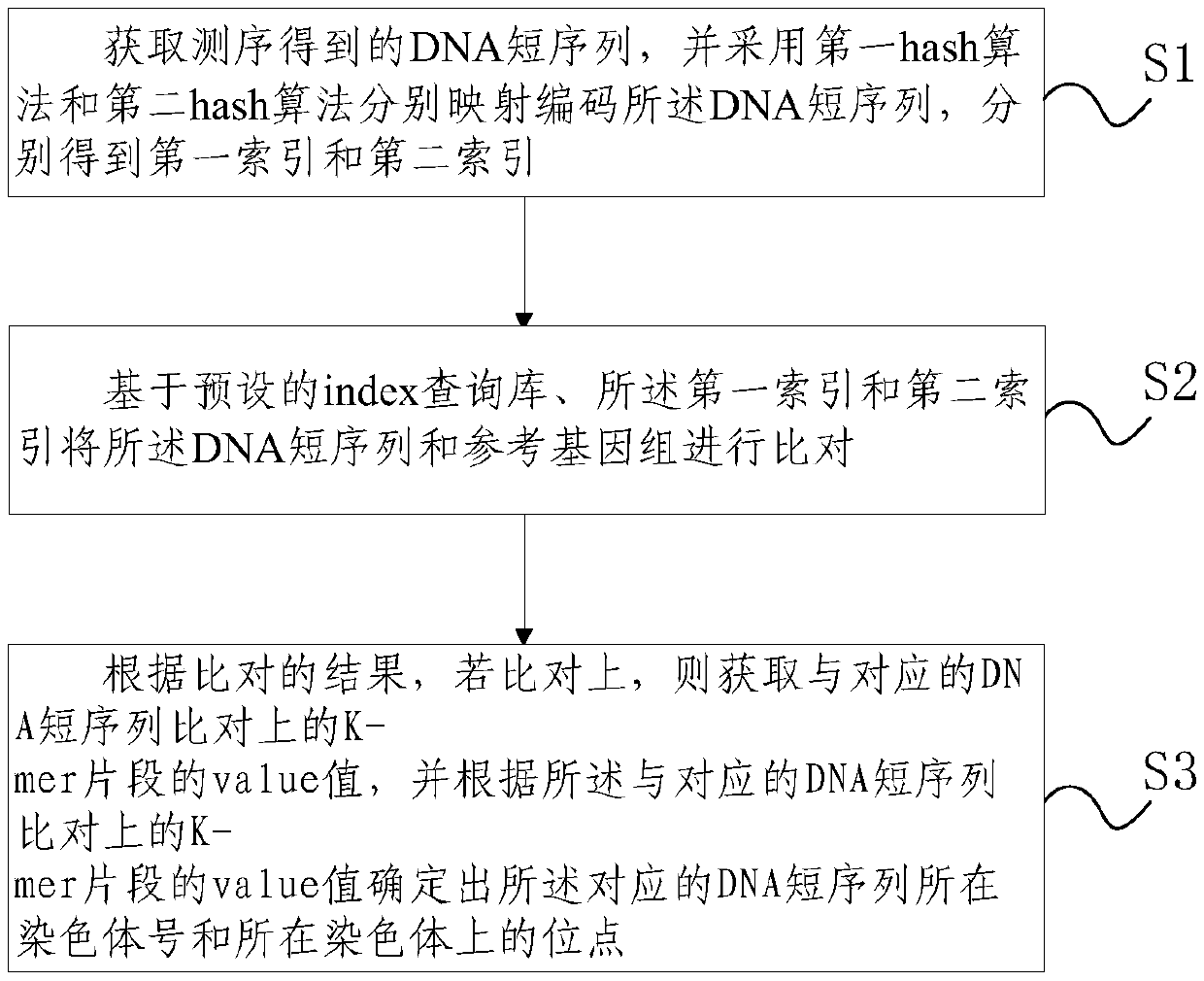
[0096] The module flow of the present invention to realize the non-invasive prenatal detection first uses perl language and shell commands to preprocess the human reference genome hg19.fa, and process it into the format required by the index query library, including three columns:
[0097] The first column: 36-mer DNA sequence fragment;
[0098] The second column: the chromosome number where the fragment is located;
[0099] The third column: the position on the chromosome where the fragment is located.
[0100] Then its core method is realized based on C language algorithm and data structure.
[0101] The design process and operation of the present invention require hardware and software environment: Linux system; more than 3 cores; more than 35 memory; C library under the Linux platform; Gcc compiler; Gdb debug...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com