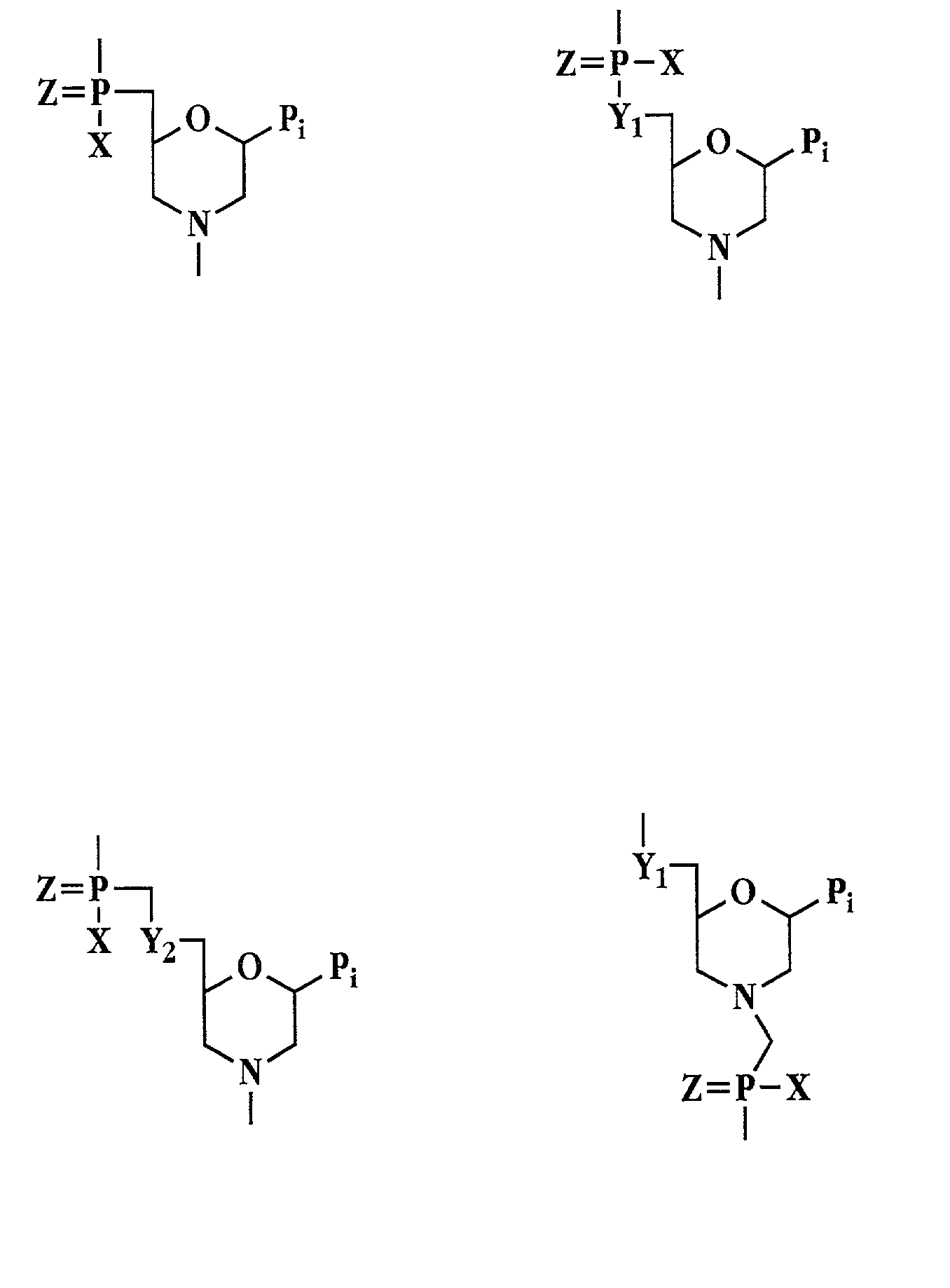
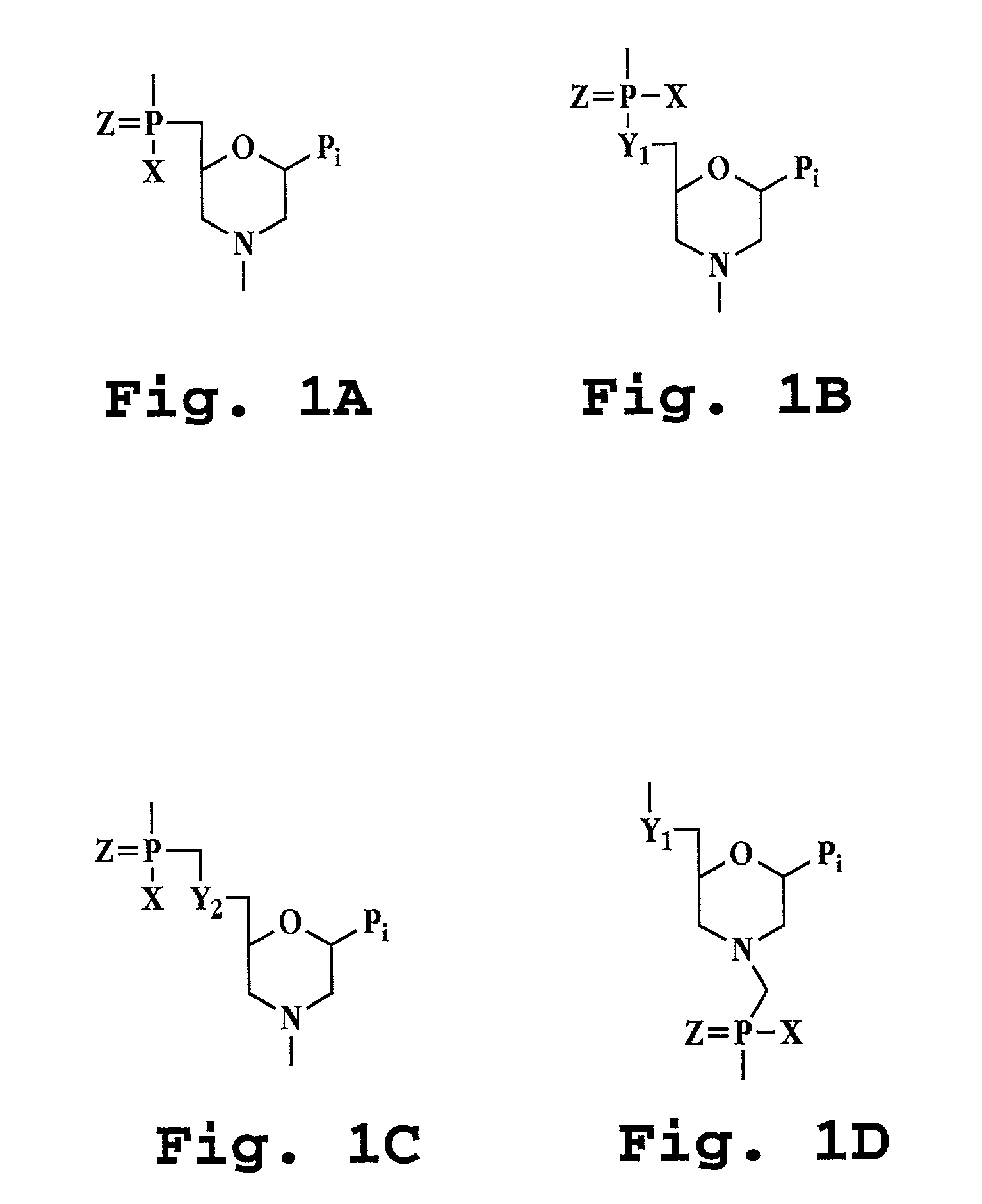
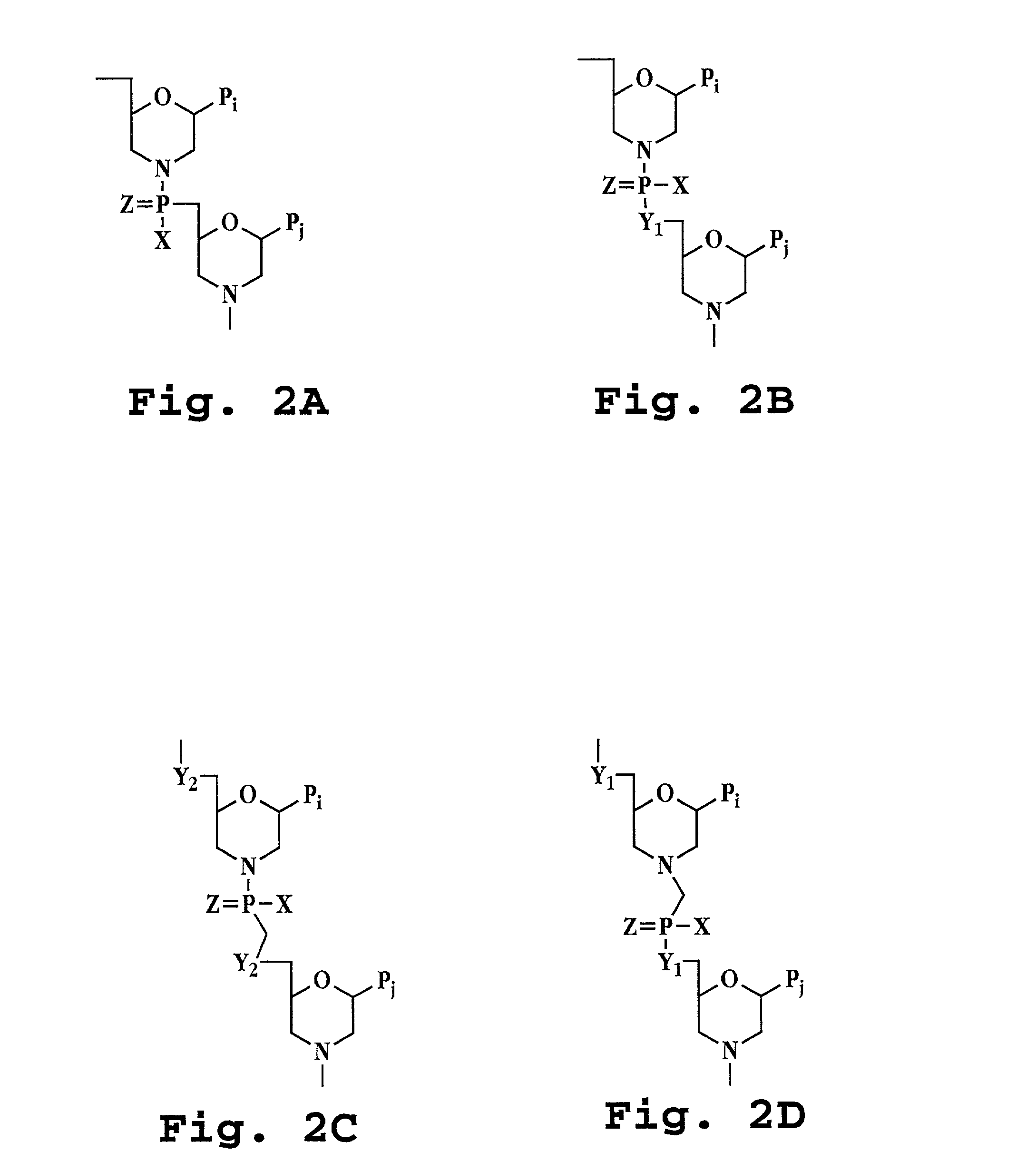
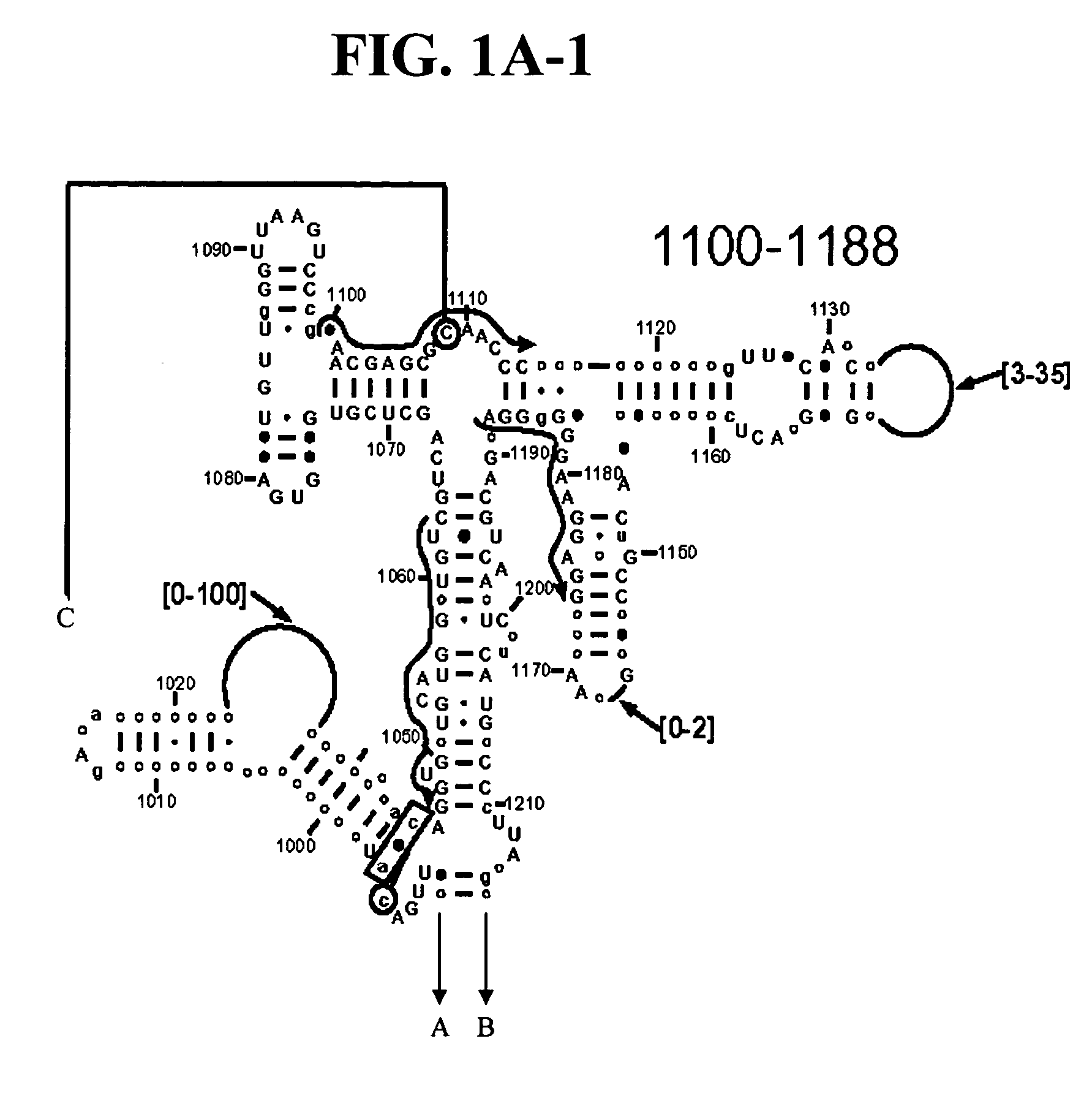
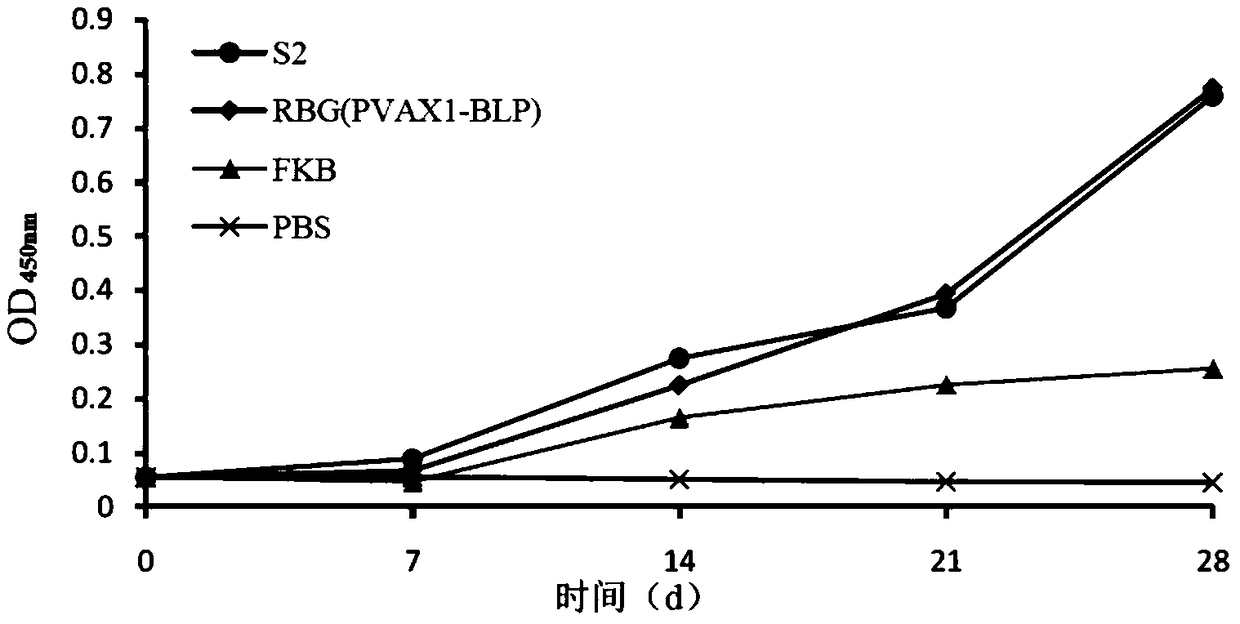
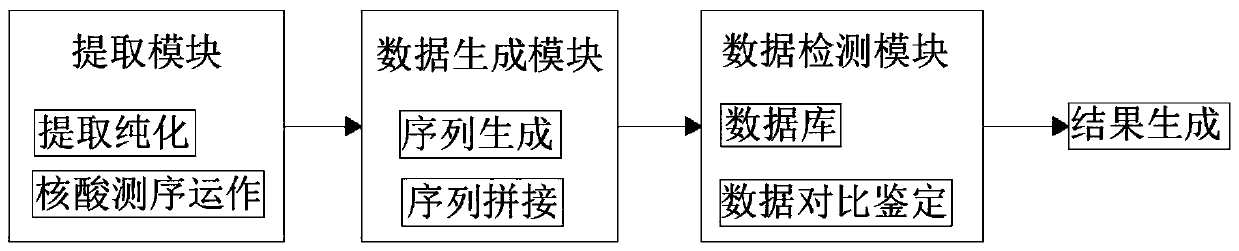
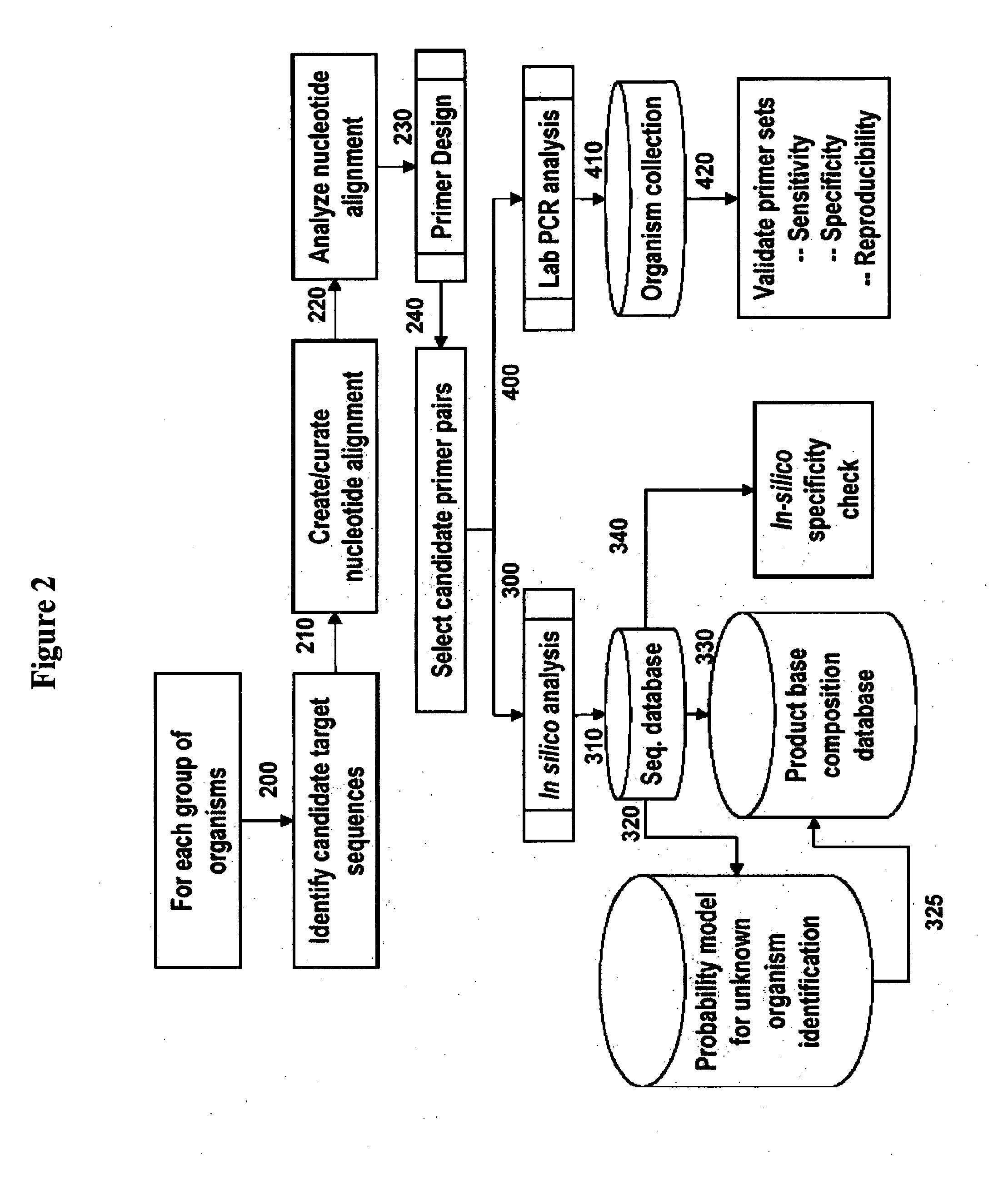
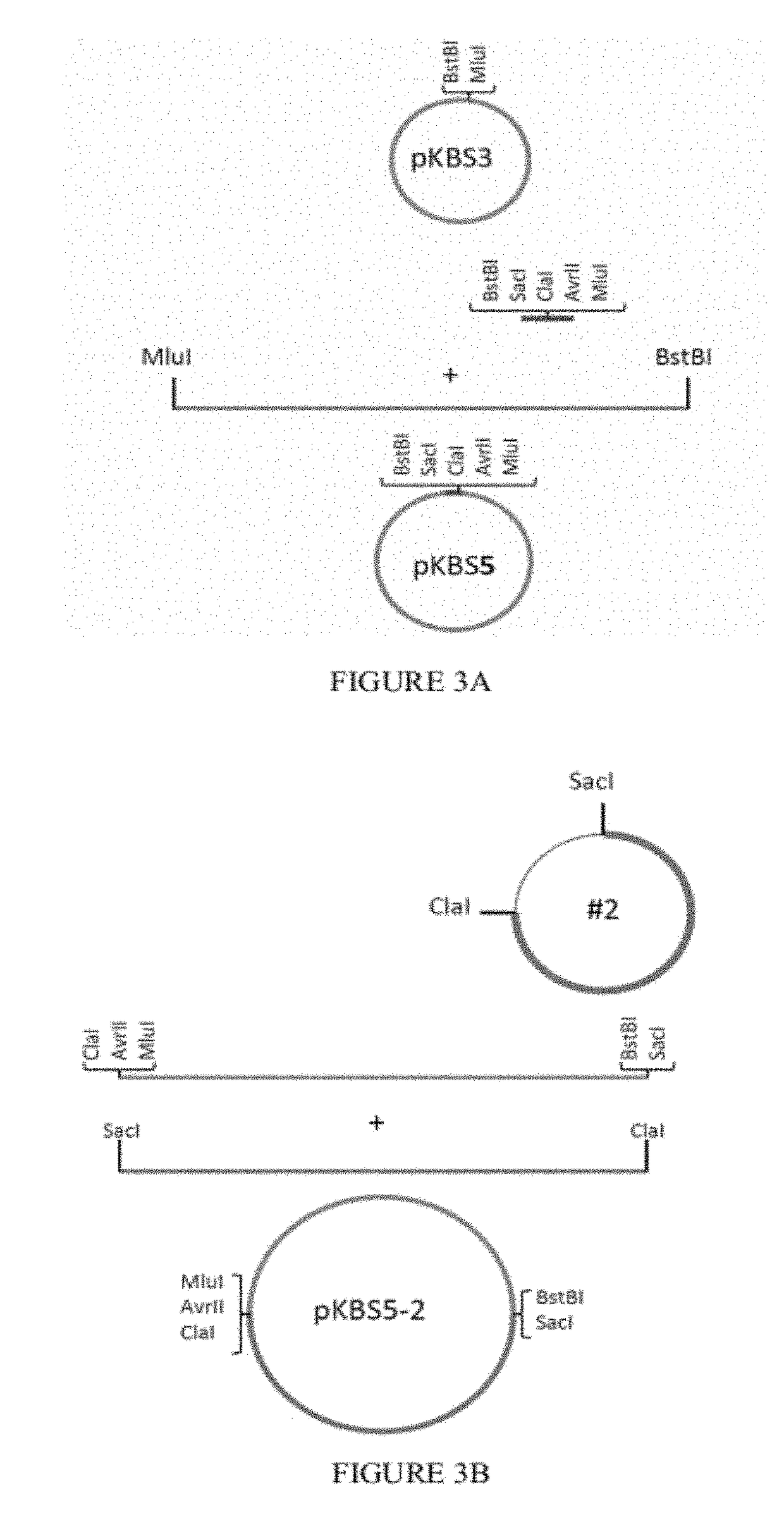
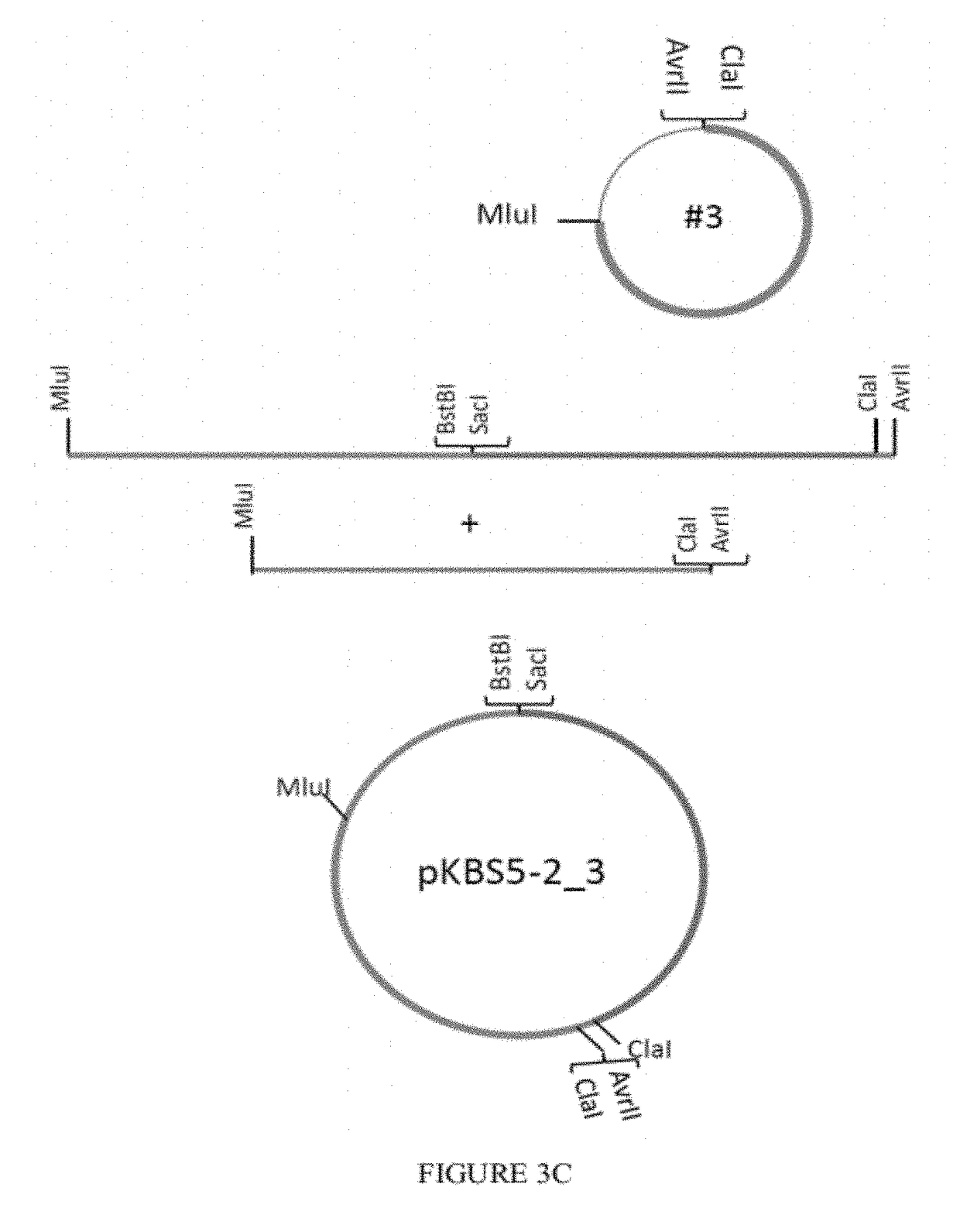
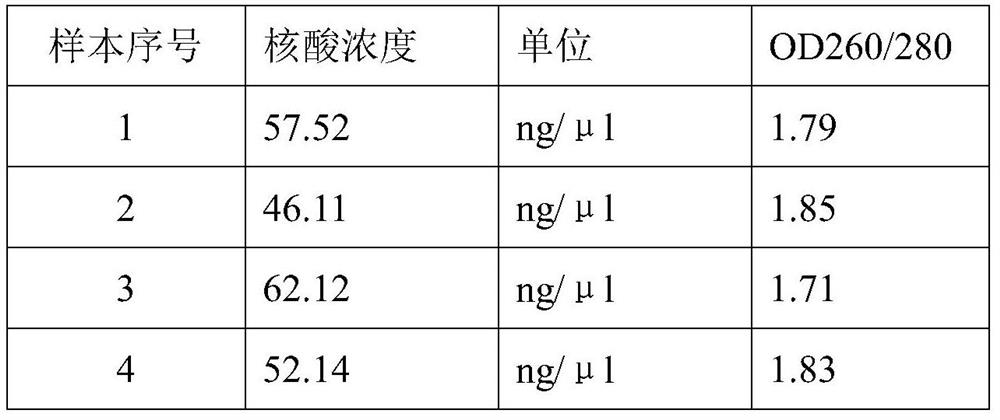
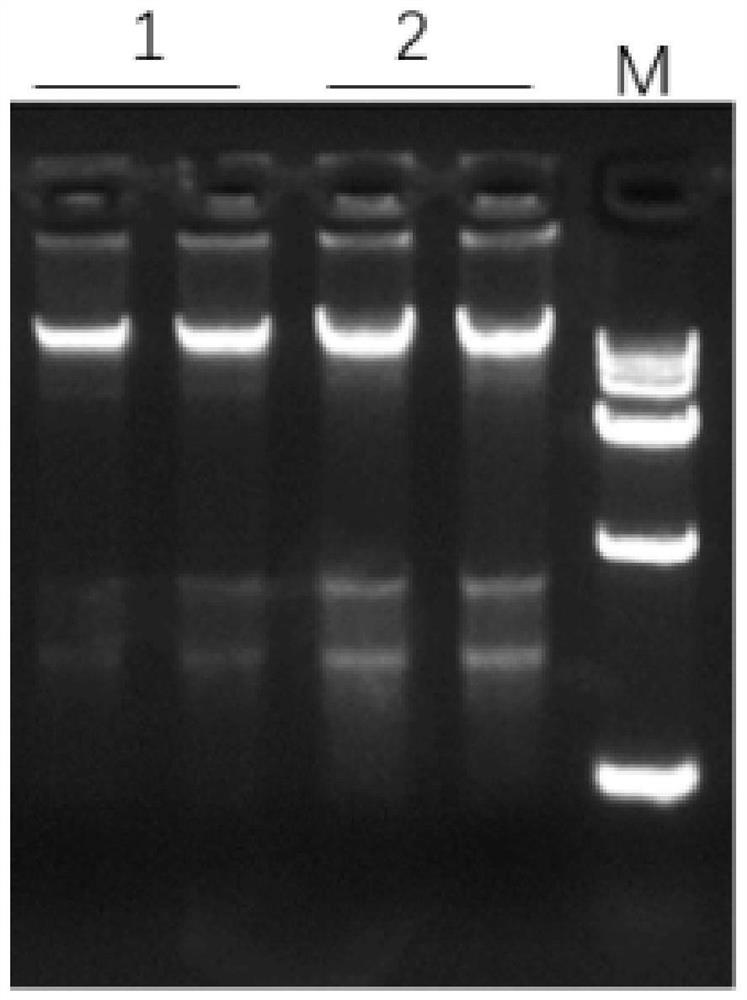
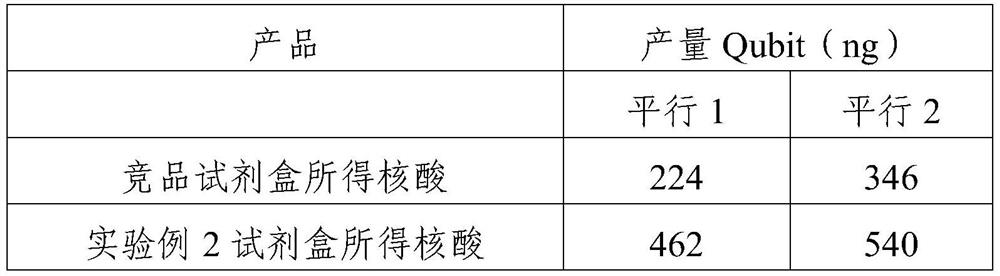
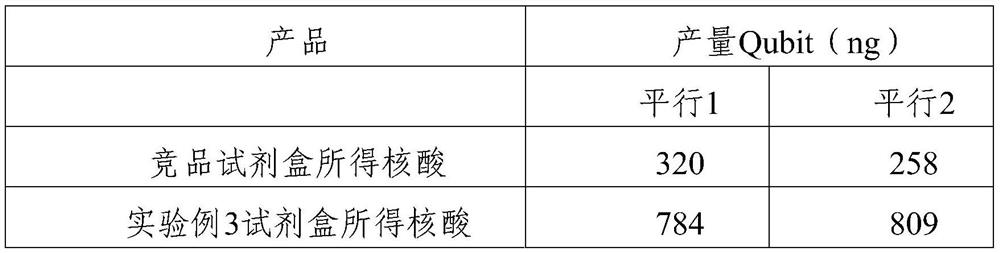
Patents
Literature
49 results about "Bacterial nucleic acid" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
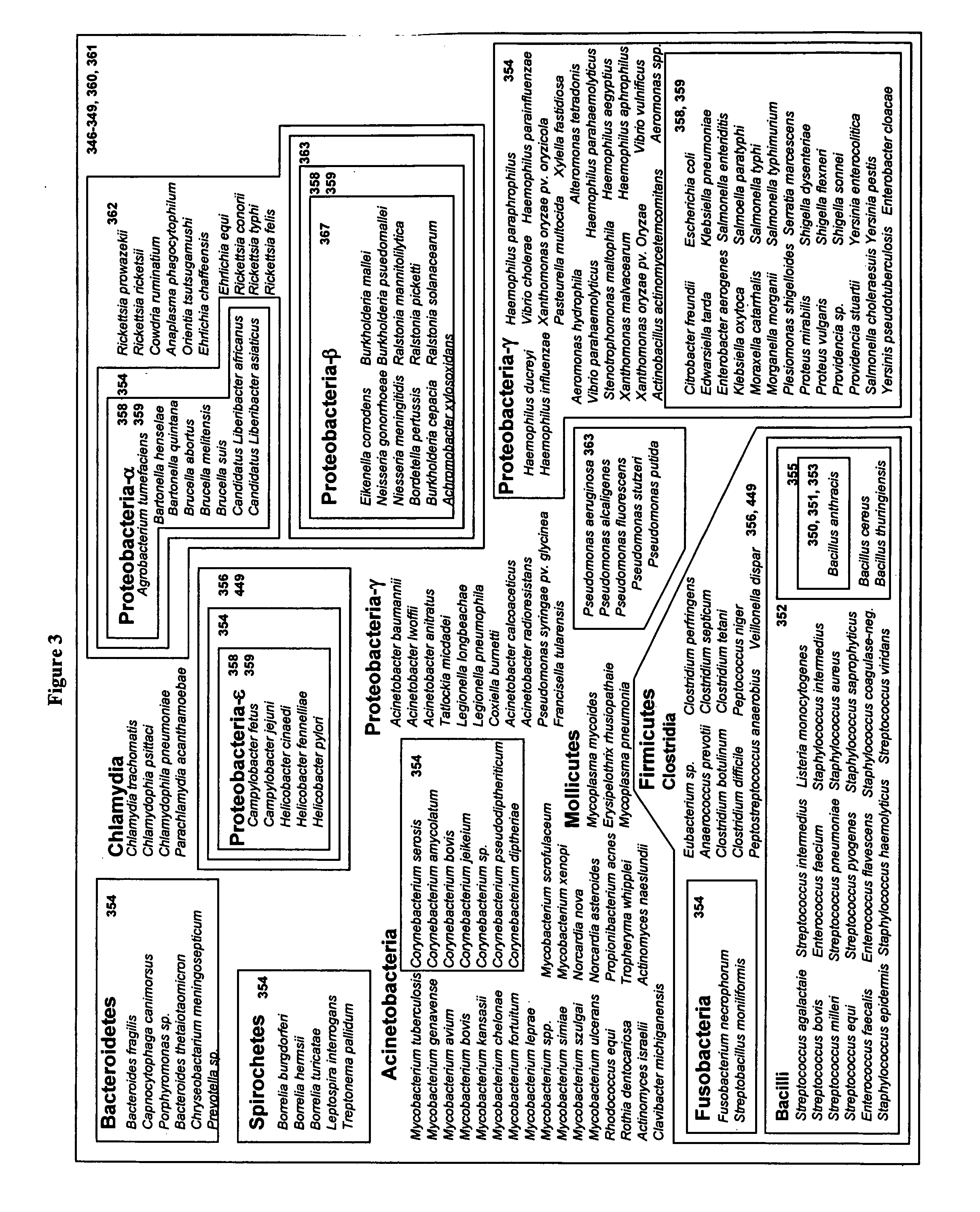
Bacteria have the same type of nucleic acids as your cells, only the arrangement is different. Bacteria have one double stranded DNA chromosome that holds rheir genetic material. Instead of linear chromosomes like we have, bacterial chromosomes are circular.
Antisense antibacterial cell division composition and method
Antisense oligomers directed to bacterial cell division and cell cycle-encoding nucleic acids are capable of selectively modulating the biological activity thereof, and are useful in treatment and prevention of bacterial infection. The antisense oligomers are substantially uncharged, and contain from 8 to 40 nucleotide subunits, including a targeting nucleic acid sequence at least 10 nucleotides in length which is effective to hybridize to (i) a bacterial tRNA or (ii) a target sequence, containing a translational start codon, within a bacterial nucleic acid which encodes a protein associated with cell division or the cell cycle. Such proteins include zipA, sulA, secA, dicA, dicB, dicC, dicF, ftsA, ftsI, ftsN, ftsK, ftsL, ftsQ, ftsW, ftsZ, murC, murD, murE, murF, murG, minC, minD, minE, mraY, mraW, mraZ, seqA, ddlB, carbamate kinase, D-ala D-ala ligase, topoisomerase, alkyl hydroperoxide reductase, thioredoxin reductase, dihydrofolate reductase, and cell wall enzyme.
Owner:SAREPTA THERAPEUTICS INC
Methods for identification of coronaviruses
InactiveUS20050266397A1SsRNA viruses positive-senseSugar derivativesMass Spectrometry-Mass SpectrometryRapid identification
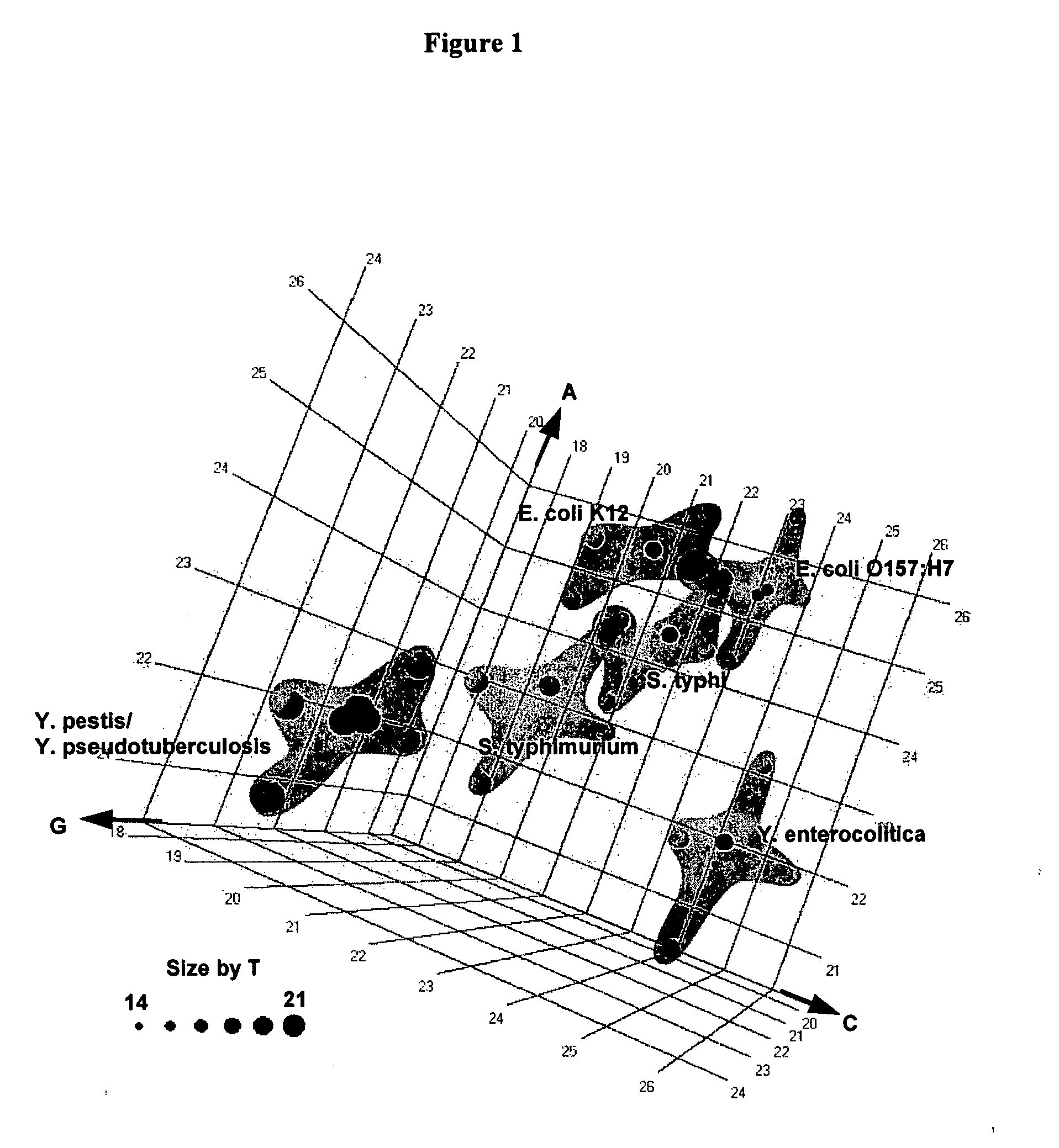
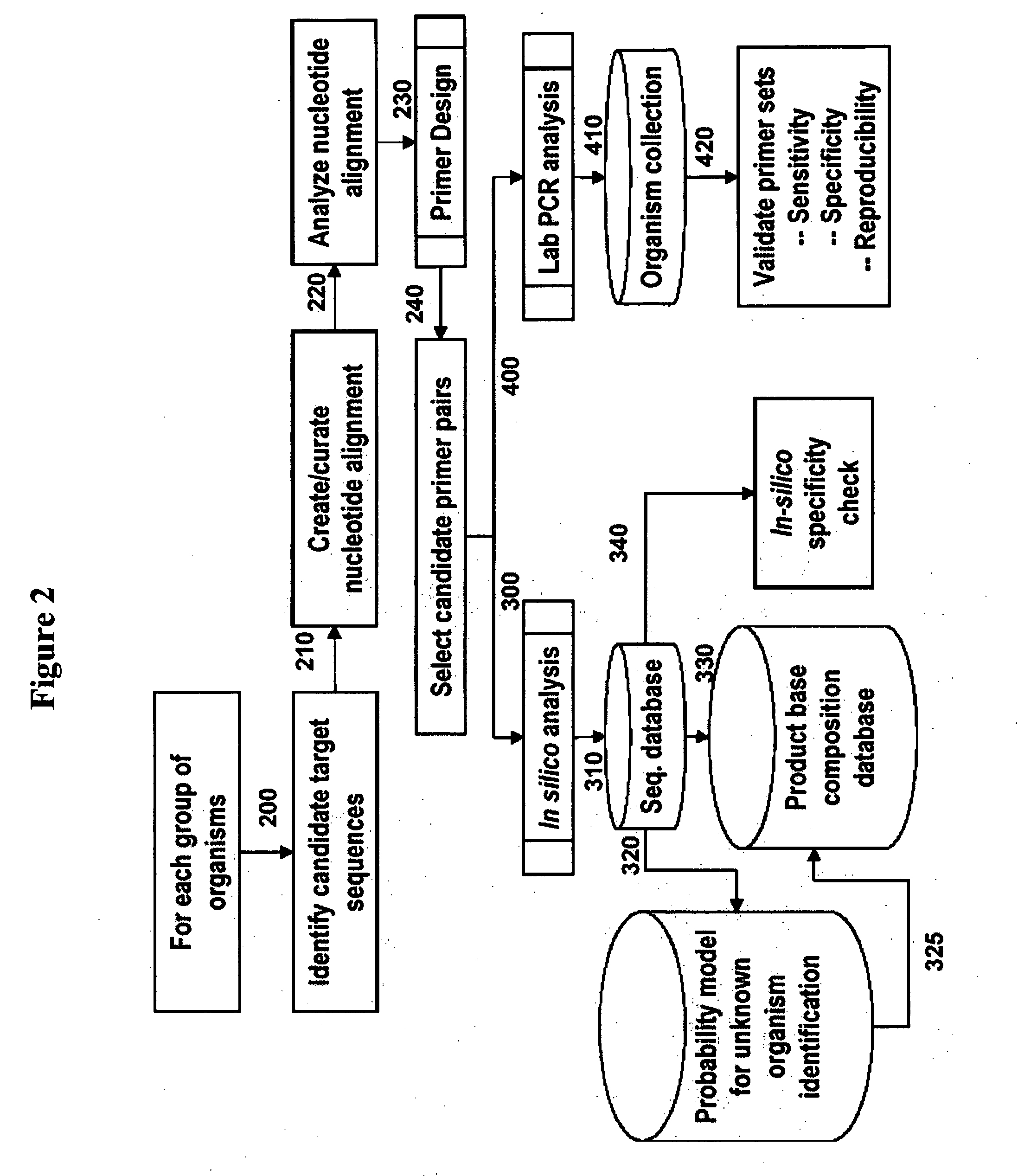
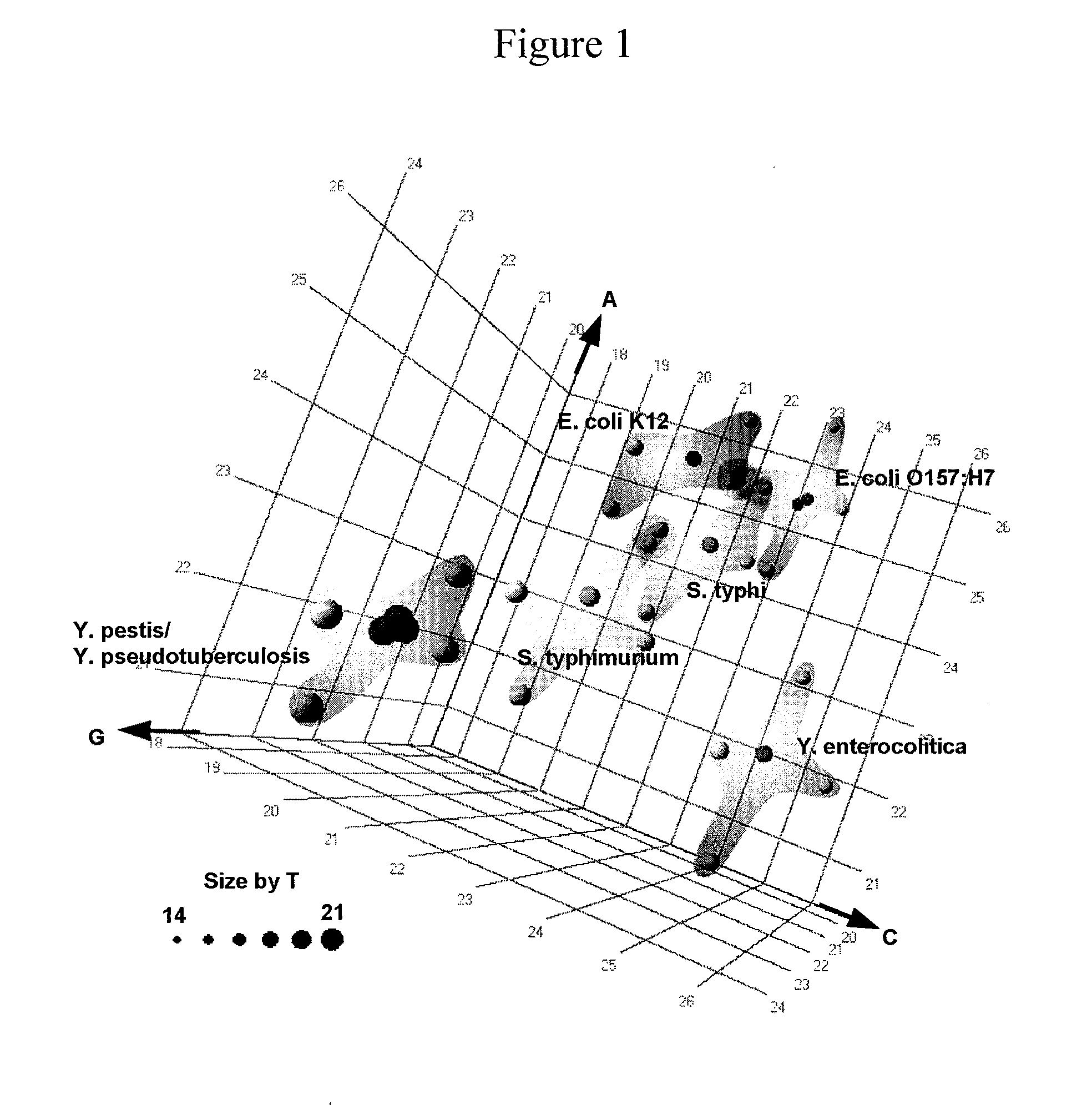
The present invention provides a method for rapid identification and quantitation of bacteria by amplification of a segment of bacterial nucleic acid followed by analysis by mass spectrometry. The compositions provide for characterization of the molecular masses and base compositions of bacterial nucleic acids which are used to rapidly identify bacteria.
Owner:IBIS BIOSCI
Process for abstracting bacterial DNA from phlegm, kit and uses thereof
ActiveCN101285062AReduce the amount of solutionEasy to operateMicrobiological testing/measurementDNA preparationWild typeSolid particle
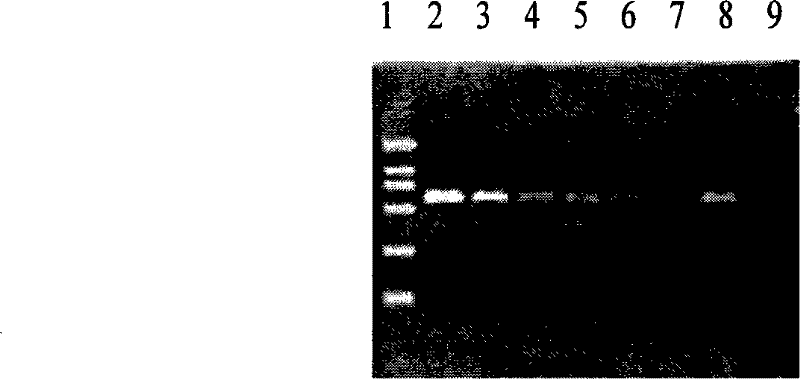
The invention discloses a method and a kit for extracting bacterial nucleic acid from sputa and applications thereof. The method is to use liquefaction reagent to liquefy a sputum sample and add solid particles to lyse cells to extract nucleic acid form the sputum. Overcoming the drawbacks of the prior method including complex procedures, a long period of time, complex operation and inadequate samples, the method of the invention for extracting nucleic acid samples is convenient and quick in operation, adequate in nucleic acid samples, low in cost and easy to implement. The method and kit for extracting bacterial nucleic acid can be used to identify strains of mycobacteria and to detect whether a gene locus of mycobacterium tuberculosis is of a wide type or a mutant type. With a chip for identifying strains and a chip for detecting whether the gene locus of mycobacterium tuberculosis is of a wide type or a mutant type, the method can identify the strains of mycobacteria and to detect whether a gene locus of mycobacterium tuberculosis is of a wide type or a mutant type in a quick, accurate and high-pass way.
Owner:CAPITALBIO CORP
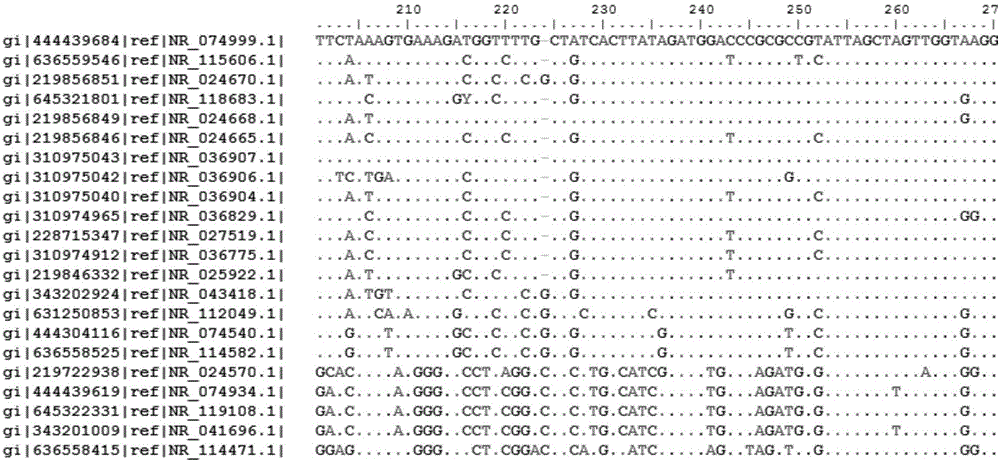
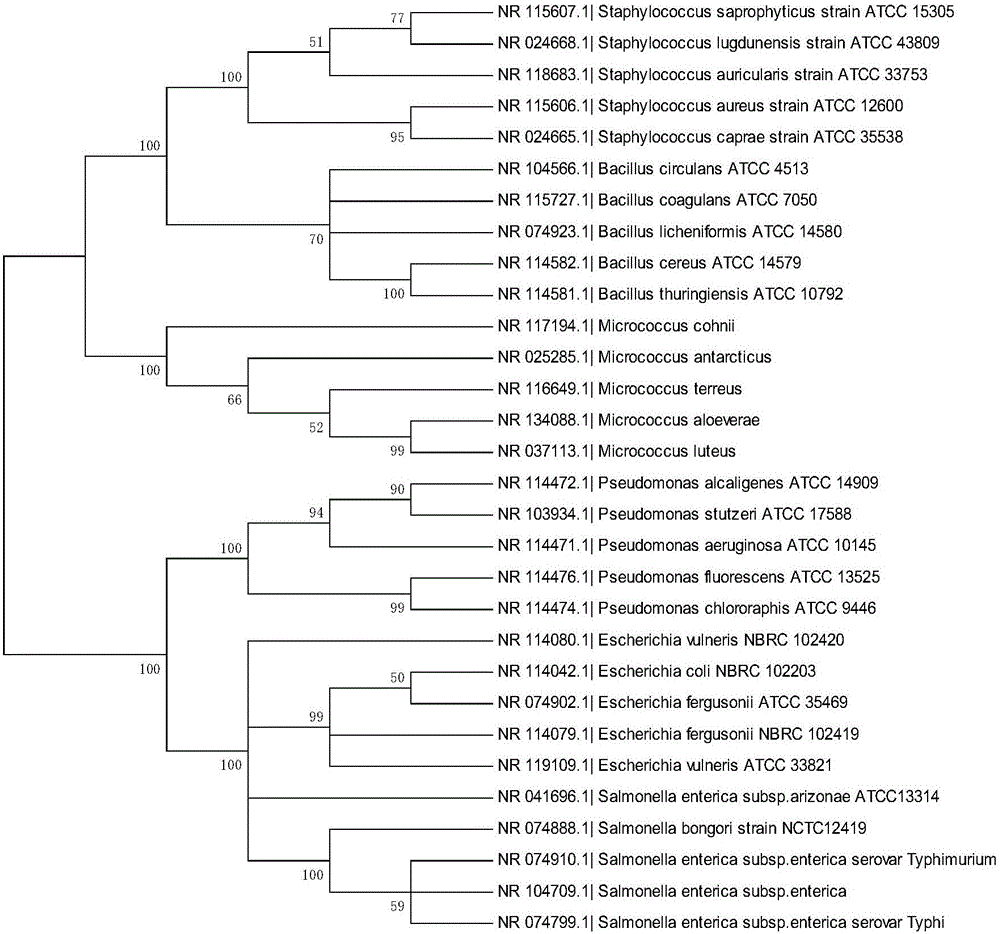
Bacterial nucleic acid sequencing identification method based on DNA bar code
InactiveCN106701914AClear scopeClear lengthMicrobiological testing/measurementSequence analysisDNA barcoding
The invention discloses a bacterial nucleic acid sequencing identification method based on a DNA bar code. Common bacterial contaminants in a medicine production environment can be effectively and accurately identified. The bacterial nucleic acid sequencing identification method comprises the steps that 1, a monoclonal bacterial strain is obtained through culture; 2, a genome DNA of the monoclonal bacterial strain obtained in the step 1 is extracted; 3, PCR is performed by using a general nucleic acid sequencing primer to amplify a specific sequence of nucleic acids; 4, a PCR product is extracted and purified; 5, a nucleic acid sequence of the PCR product obtained in the step 4 is determined; 6, a sequence analysis software is applied to perform sequence splicing, positioning is performed by using a general nucleic acid sequencing primer pair, a primer region is removed, and a DNA sequence of a corresponding fragment is obtained; 7, the DNA sequence of the corresponding fragment in the step is imported into a DNA bar code standard sequence core database or a GenBank database for sequence comparison, and bacteria are identified.
Owner:SHANGHAI INST FOR FOOD & DRUG CONTROL
Compositions for use in identification of bacteria
InactiveUS20100035239A1Microbiological testing/measurementOligonucleotide primersRapid identification
The present invention provides oligonucleotide primers and compositions and kits containing the same for rapid identification of bacteria by amplification of a segment of bacterial nucleic acid followed by molecular mass analysis.
Owner:IBIS BIOSCI
Method and reagent for extraction of viral/bacterial nucleic acid in animal sample
ActiveCN109385418AAvoid cloggingEasy to operateMicrobiological testing/measurementDNA preparationSilica gelDigestion
The invention discloses a method and a reagent for extraction of viral / bacterial nucleic acid in an animal sample. The method comprises the following steps: taking the animal sample, and adding a lysate for lysis at a room temperature so as to obtain a lysis reaction solution; and adding an ethanol solution into the lysis reaction solution, transferring an obtained mixture into a silica gel containing adsorption membrane centrifugal column, carrying out adsorption with a manner of low-speed centrifugation, and carrying out washing and eluting. According to the invention, the bacterial nucleicacid extracted by using the method provided by the invention has high purity; no proteinase K for sample digestion is needed to be used in the process of extraction; reagent components and an operation flow are simplified; the cost of hardware equipment can be reduced through a manner of low-speed centrifugation in the process of extraction; a low-speed palm centrifuge can be adopted, is convenient to carry and is applicable to on-site operation; the components of the lysate are optimized; the binding ability of nucleic acid molecules to a silica-gel adsorption membrane is reinforced; and goodnucleic acid extraction efficiency is guaranteed.
Owner:USTAR BIOTECHNOLOGIES (HANGZHOU) CO LTD
Method for preparing bacterial nucleic acid fingerprint characteristic spectrum library
ActiveCN103352257AValid identificationUndisturbedMicrobiological testing/measurementMicroorganism based processesEnzyme digestionDigestion
The invention discloses a method for preparing a bacterial nucleic acid finger print library, wherein the method comprises PCR amplification, SAP enzyme digestion, transcription, nucleic acid digestion through enzyme, purification, mass spectrometer detection and other steps. According to the present invention, based on the method, a nucleic acid finger print library of the common bacterial is established; and according to a mass spectrum characteristic spectrum library produced by experiments, classification and identification can be performed on bacterial to be detected, and results can be widely used for bacterial classification and identification, phylogenetic analysis, drug screening, import and export inspection and other fields.
Owner:BIOYONG TECH
Bacteriophage genome DNA (deoxyribonucleic acid) extraction kit and method
ActiveCN105567678AMeet the needs of molecular experiments such as sequencingEliminate Interference ProblemsDNA preparationProtein structureDNA Contamination
The invention provides a bacteriophage genome DNA (deoxyribonucleic acid) extraction kit and method. The kit comprises a reagent A (chloroform), a reagent B (DNase I), a reagent C (RNase A), a reagent D (precipitation buffer solution), a reagent E (pyrolysis buffer solution), a reagent F (primary pyrolysis main solution), a reagent G (secondary pyrolysis solution), a reagent H (impurity removal solution), a reagent I (DNA combination buffer solution), a reagent J (rinsing buffer solution) and a reagent K (DNA eluate). The extraction method comprises the following steps: removal of bacterium cells and fragments, degradation of bacterium nucleic acid, bacteriophage particle precipitation, bacteriophage capsid protein structure damage and hydrolysis, impurity removal, DNA liquid separation, DNA adsorption, cleaning of adsorbed DNA and elution of adsorbed DNA. The kit and method can be widely used for extracting the bacteriophage genome DNA, and has the advantages of high extraction speed, high extracted DNA yield and high purity, and the extracted DNA can not be polluted by the host bacterium genome DNA.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Compositions and methods for detecting BV-associated bacterial nucleic acid
ActiveUS20140302500A1Opens up structureStrong specificityMicrobiological testing/measurementBacterial vaginosisOligomer
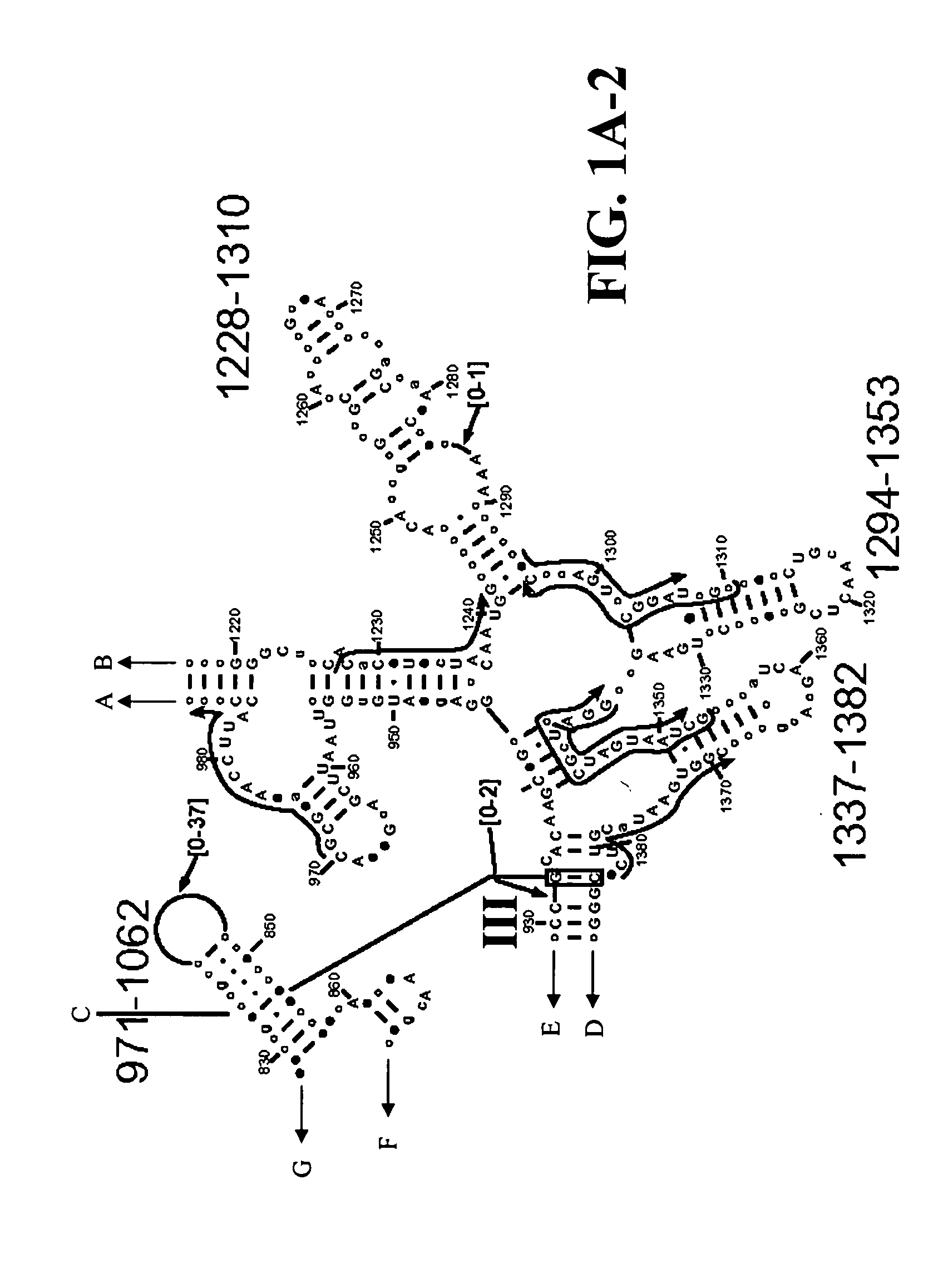
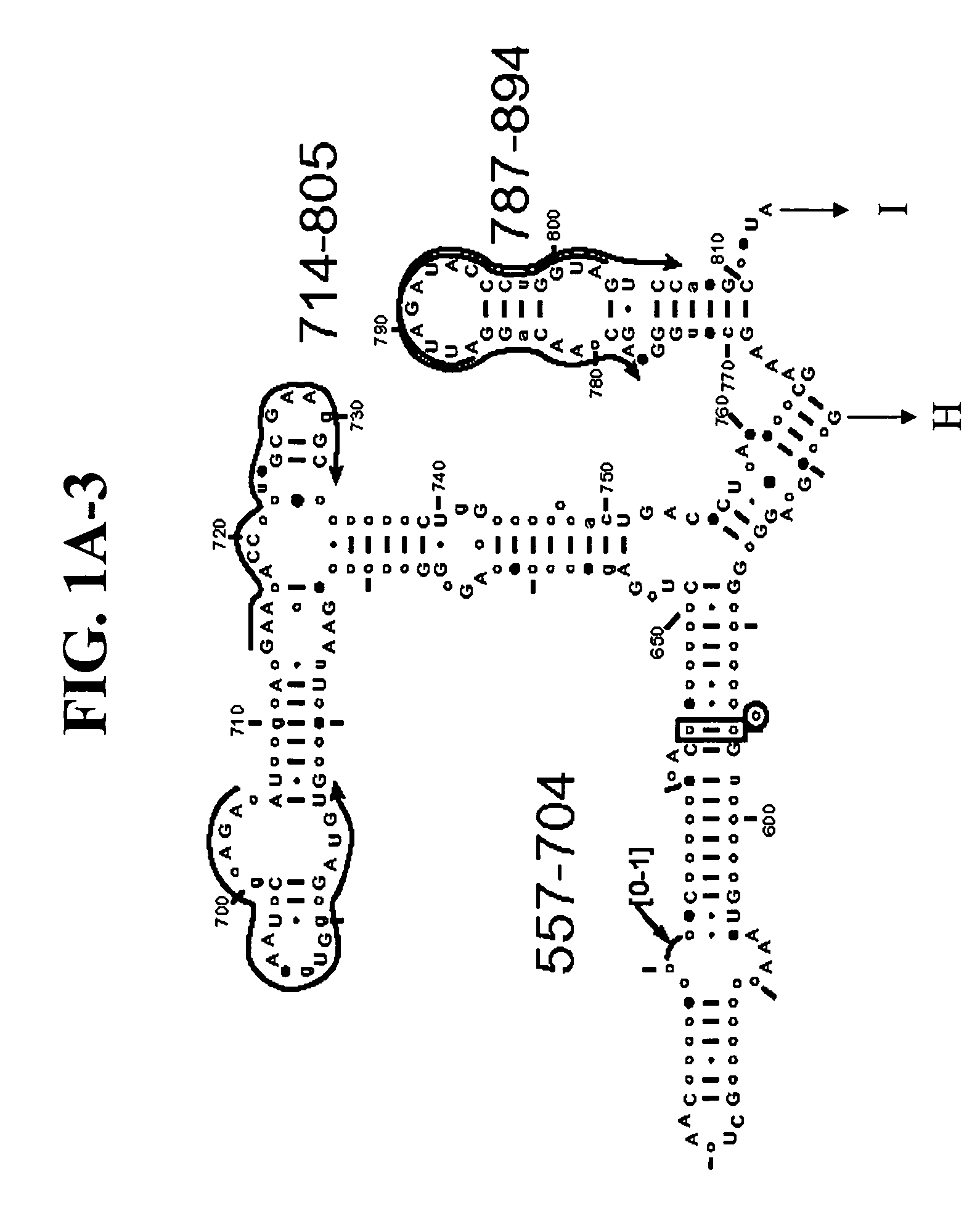
Disclosed are nucleic acid oligomers, including amplification oligomers, capture probes, and detection probes, for detection of a 16S rRNA or its encoding gene from bacterial species associated with bacterial vaginosis. Also disclosed are methods of specific nucleic acid amplification and detection using the disclosed oligomers, as well as corresponding reaction mixtures and kits.
Owner:GEN PROBE INC
DNA loaded Brucella ghost composite vaccine
InactiveCN108690823ALittle side effectsImprove securityAntibacterial agentsBacterial antigen ingredientsSide effectA-DNA
The invention discloses a DNA loaded Brucella ghost composite vaccine. The preparation method comprises following steps: introducing a suicide plasmid that contains a nucleic acid molecule encoding atemperature sensitive regulatory protein cI857, a nucleic acid molecule encoding a bacteriophage splitting protein E, and a nucleic acid molecule encoding a bacterial nuclease protein A into Brucella;utilizing a homologous recombination technology to obtain recombinant Brucella; culturing the recombinant Brucella to obtain a bacterial solution, processing the bacterial solution at a high temperature, collecting bacterial cells, and adding target DNA to obtain the DNA loaded Brucella ghost composite vaccine. The composite vaccine has following advantages: (1) the vaccine has the characteristics of bacterial ghost, compared with a conventional killed vaccine or an attenuated live vaccine, the side effect of the composite vaccine is small, the safety is high, and the protection effect is good; and (2) the bacterial ghost is a safe and effective carrier for delivering DNA vaccines, can introduce nucleic acid vaccines into antigen presenting cells, and performs high efficient expression. The composite vaccine has an important meaning for controlling the epidemic spreading of brucellosis and has a wide application range.
Owner:INNER MONGOLIA HUAXI BIOTECH
Antisense antibacterial cell division composition and method
Antisense oligomers directed to bacterial cell division and cell cycle-encoding nucleic acids are capable of selectively modulating the biological activity thereof, and are useful in treatment and prevention of bacterial infection. The antisense oligomers are substantially uncharged, and contain from 8 to 40 nucleotide subunits, including a targeting nucleic acid sequence at least 10 nucleotides in length which is effective to hybridize to (i) a bacterial tRNA or (ii) a target sequence, containing a translational start codon, within a bacterial nucleic acid which encodes a protein associated with cell division or the cell cycle. Such proteins include zipA, sulA, secA, dicA, dicB, diCc, dicF, ftsA, ftsl,ftsN, ftsK, ftsL, ftsQ, ftsw, ftsZ, murC, murD, murE, murF, murg, minC, minD, minE, mraY, mraW, mraZ, seqA, ddlB, carbamate kinase, D-ala D-ala ligase, topoisomerase, alkyl hydroperoxide reductase, thioredoxin reductase, dihydrofolate reductase, and cell wall enzyme.
Owner:SAREPTA THERAPEUTICS INC
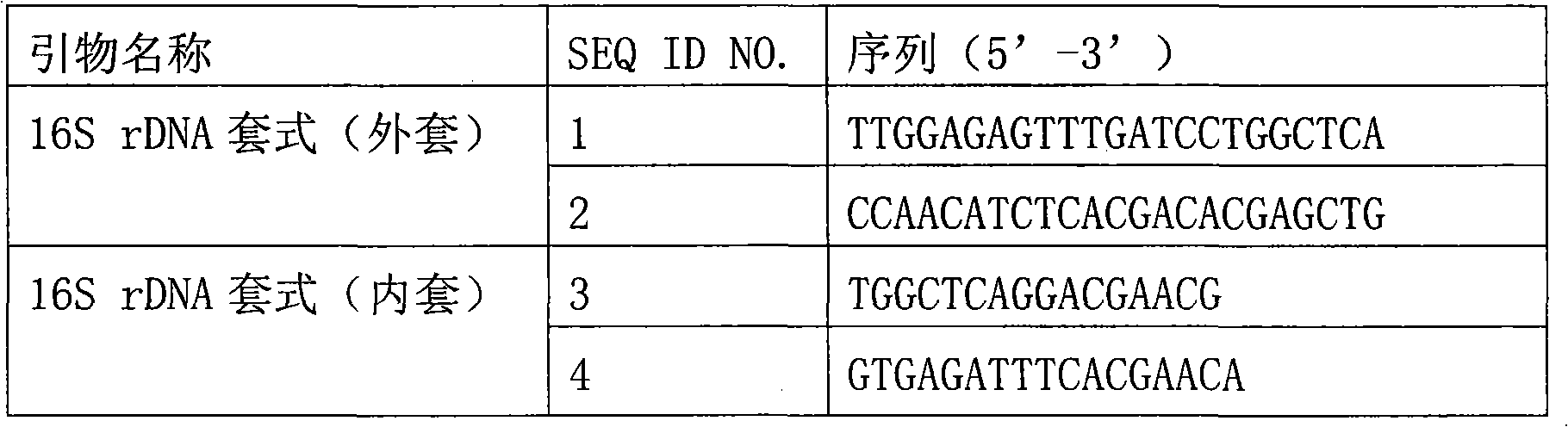
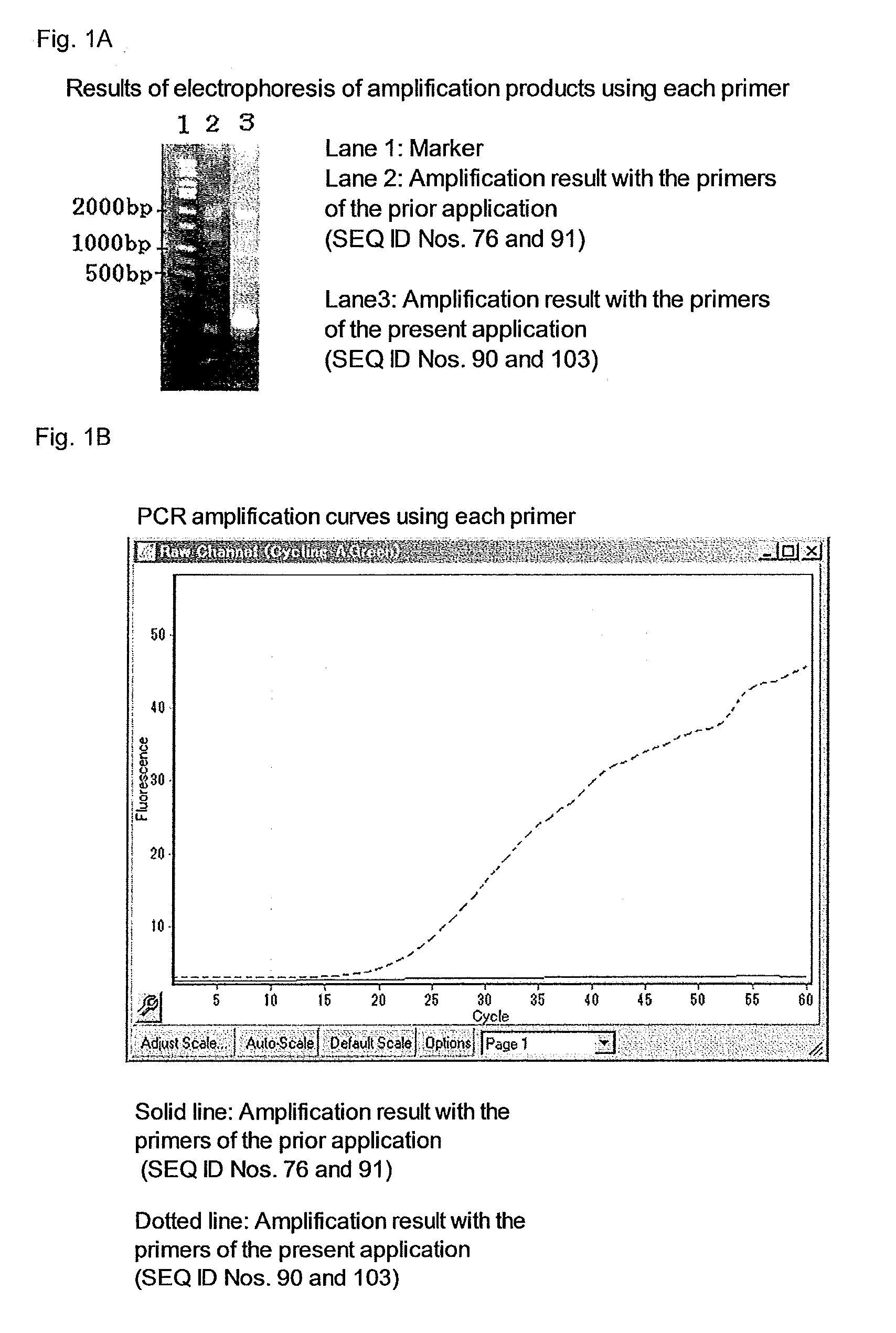
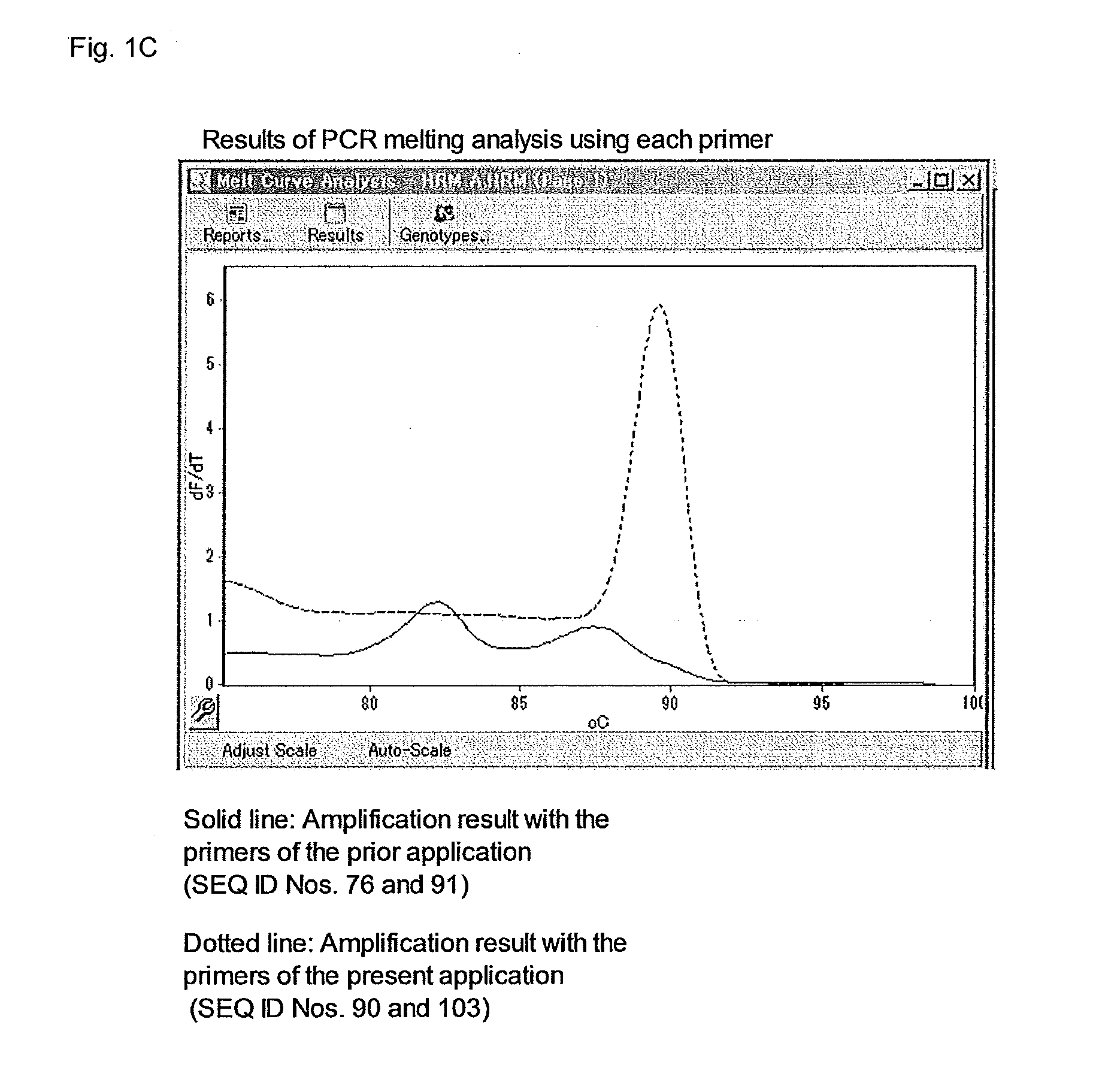
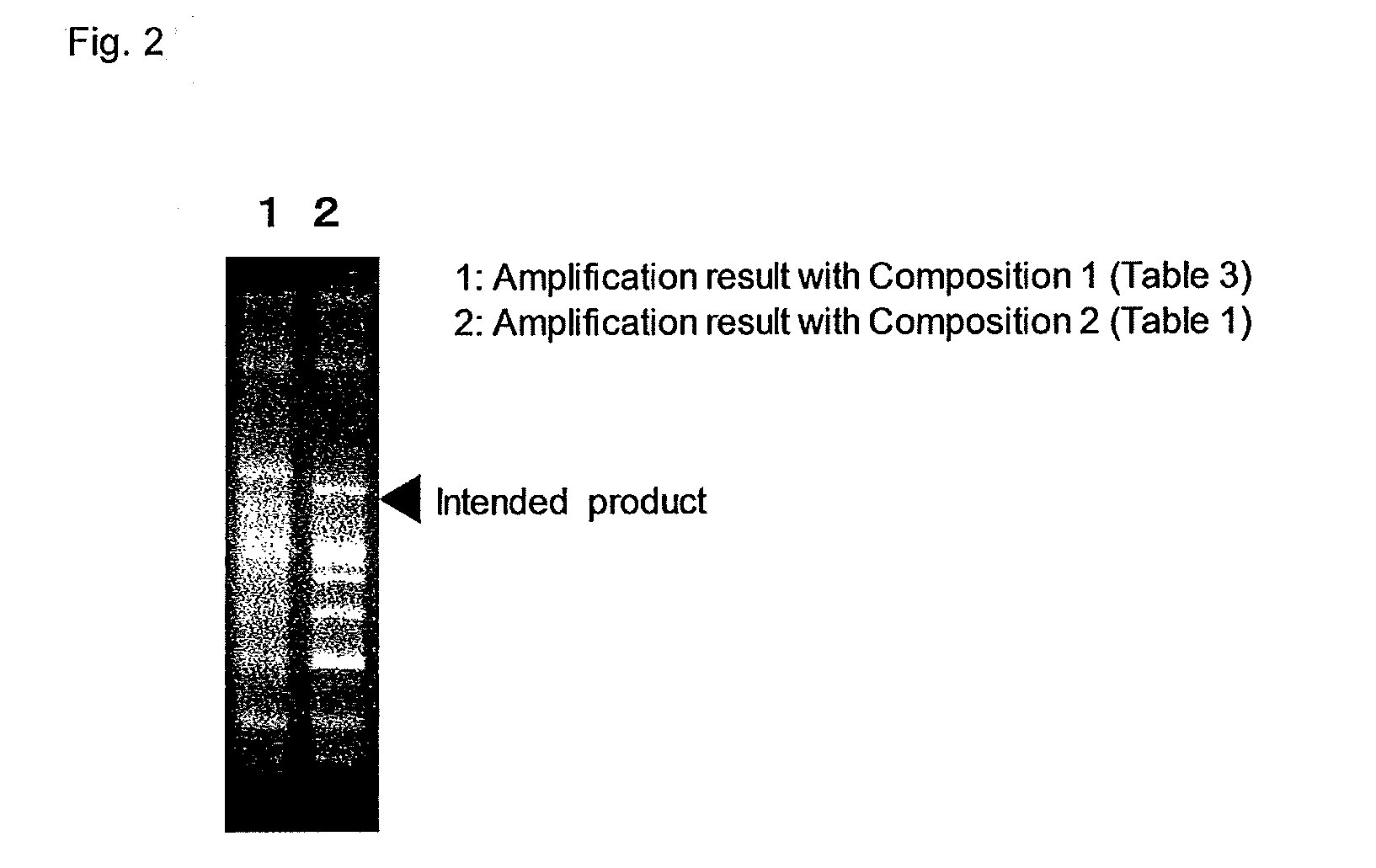
Primer set for PCR for bacterial DNA amplification, kit for detection and/or identification of bacterial species, and, method of detection and/or identification of bacterial species
InactiveUS20160244812A1High sensitivityEasy to detectMicrobiological testing/measurementPcr methodMicrobiology
A primer set useful for further improvement of sensitivity in detection or identification of bacterial species in a sample by a PCR method, a kit for PCR using the primer set and a method of detection or identification of bacterial species in a sample using the primer set. Sensitivity in detection or identification of bacterial species in a sample by conducting a PCR method using the primer set with a minimized contamination amount of bacterial nucleic acid is improved. The primer set includes at least one primer pair in seven primer pairs obtained by selecting one primer pair from each of Groups S1 to S7 consisting of specific primers. In addition, the kit for detection of bacterial species includes at least one primer pair in seven primer pairs obtained by selecting one primer pair from each of Groups K1 to K7 consisting of specific primers.
Owner:MITSUI CHEM INC
Intestinal flora nucleic acid extraction system
PendingCN110437970ABioreactor/fermenter combinationsBiological substance pretreatmentsCentrifugationGut flora
The invention discloses an intestinal flora nucleic acid extraction system, and belongs to the technical field of medical clinical examination. The system can mainly solve problems that existing traditional methods are time-consuming and labor-consuming and low in working efficiency and experiment results and quality cannot be guaranteed. The main characteristics of the system are that the systemincludes a specimen sample introduction module, an intelligent centrifugation module, a test tube sample introduction module, a mechanical arm module and a reagent sample introduction module; the intelligent centrifugation module is located in the middle of a machine base plate; the specimen sample introduction module and the reagent sample introduction module are arranged on the middle layer frame of a machine frame and located at the upper side part of the intelligent centrifugation module; the reagent sample introduction module is movably arranged on the middle layer frame of the machine frame and located above the intelligent centrifugation module; and the mechanical arm module is arranged on the upper layer frame of the machine frame. The system is mainly used for automatically extracting bacterial nucleic acid in manure.
Owner:湖北微伞医疗科技有限公司
16S rDNA based preparation method of bacteria nucleic acid fingerprint characteristic spectrums and application thereof
InactiveCN103060431AValid identificationUndisturbedMicrobiological testing/measurementMicroorganism based processesEnzymatic digestionSuper absorbent
The invention discloses a method for preparing bacteria 16S rDNA fingerprint spectrums. The method comprises the steps of PCR (polymerase chain reaction) amplification, SAP (super absorbent polymer) enzymatic digestion, transcription and nuclease digestion, purification, mass spectrometer detection and the like. A nucleic acid fingerprint spectrum database of common bacteria is created based on the method. According to the mass spectrum peak chart generated through experiments, the bacteria to be detected can be classified and identified and the results can be widely applied to the fields of bacteria classification and identification, genetic evolution analysis, drug resistance screening application, import and export inspection and the like.
Owner:向华
Nested PCR detection method or/and identification method of brucella
InactiveCN109306372AHigh sensitivityStrong specificityMicrobiological testing/measurementMicroorganism based processesTissue fluidBrucella species
The invention relates to a nested PCR detection method or / and an identification method of brucella, and relates to the technical field of molecular biology. The nested PCR technique of the invention can not only detect and identify pure bacterial nucleic acid DNA, but also detect and identify trace brucella nucleic acid DNA of tissue fluid, blood, and the like; and can not only identify brucella species of the nucleic acid DNA of the sample to be tested, but also identify the biotype thereof. The nested PCR result analysis can be standardized, with simple test operation, and can be completed by a person with ordinary PCR techniques.
Owner:ICDC CHINA CDC
Bacterial nucleic acid sequencing identification method and bacterial identification kit based on DNA characteristic sequence
PendingCN108410971AReliable Judgment BasisRapid Integrated IdentificationBioreactor/fermenter combinationsBiological substance pretreatmentsBacteria identificationGenomic DNA
The invention relates to a bacterial nucleic acid sequencing identification method and kit based on DNA characteristic sequence. The method comprises the steps: selecting a bacterial strain to be identified, and extracting and purifying a genomic DNA; amplifying a characteristic sequence fragment of the purified genomic DNA by PCR; extracting and purifying the PCR amplified product; determining asequence of the purified PCR amplified product, and removing a primer region by universal sequence splicing software, to obtain a DNA sequence of the corresponding fragment; importing the DNA sequenceof the corresponding fragment into a standard nucleic acid sequence database, carrying out sequence comparison, and carrying out bacterial strain identification, wherein the extraction and purification of the genomic DNA of the bacterial strain adopts lysozyme, sodium dodecyl sulfate and cetyltriethylammnonium bromide for bacterial crushing; nucleic acid is precipitated by phenol-chloroform-isoamyl alcohol and ethanol to obtain the genomic DNA which meets the quality control requirements. An objective and accurate judgment basis can be provided for the bacterial nucleic acid sequencing identification method, and rapid and integrated identification of bacterial pollutants is achieved.
Owner:SHANGHAI INST FOR FOOD & DRUG CONTROL
Enzyme digestion based preparation method of bacteria nucleic acid fingerprint characteristic spectrums and application thereof
InactiveCN103060926AValid identificationUndisturbedMicrobiological testing/measurementLibrary creationEnzymatic digestionSuper absorbent
The invention discloses a method for preparing bacteria nucleic acid fingerprint spectrums. The method comprises the steps of PCR (polymerase chain reaction) amplification, SAP (super absorbent polymer) enzymatic digestion, transcription and nuclease digestion, purification, mass spectrometer detection and the like. A nucleic acid fingerprint spectrum database of common bacteria is created based on the method. According to the mass spectrum peak chart generated through experiments, the bacteria to be detected can be classified and identified and the results can be widely applied to the fields of bacteria classification and identification, genetic evolution analysis, drug resistance screening application, import and export inspection and the like.
Owner:BIOYONG TECH
Compositions and methods for detecting bv-associated bacterial nucleic acid
ActiveUS20140065617A1Opens up structureStrong specificitySugar derivativesMicrobiological testing/measurementBacterial vaginosisOligomer
Disclosed are nucleic acid oligomers, including amplification oligomers, capture probes, and detection probes, for detection of a 16S rRNA or its encoding gene from bacterial species associated with bacterial vaginosis. Also disclosed are methods of specific nucleic acid amplification and detection using the disclosed oligomers, as well as corresponding reaction mixtures and kits.
Owner:GEN PROBE INC
Bacterial nucleic acid sequencing and identification system and method for intelligent DNA barcodes
InactiveCN109722484AConvenient sequencingEasy to identifyMicrobiological testing/measurementFood poisoningA-DNA
The invention discloses a bacterial nucleic acid sequencing and identification system and method for intelligent DNA barcodes, and relates to the technical field of DNA sequencing and identification.Through the system and method, technical research on the DNA barcode database and detection and identification traceability thereof of food poisoning sources can be conveniently realized, so that foodsafety can be guaranteed, the meal selection of people can be guided, the occurrence of poisoning events can be prevented, and as the nucleic acid sequencing identification method of common bacteriais established, convenient DNA barcode sequencing and identification of bacteria, cells and viruses in the biological field can be achieved; operation can be performed on a DNA genome in a special wayduring extraction and purification, so that the dissociation of a DNA sequence can be avoided. Through the adoption of a constructed standard nucleic acid sequence, a standard nucleic acid sequence database for bacterial nucleic acid sequencing and identification can be input according to a unified common description and normative codes, so that objective and accurate judgment bases can be provided for the nucleic acid sequencing identification method.
Owner:青岛市疾病预防控制中心
Composition For Use In Identification Of Bacteria
InactiveUS20120171692A1Sugar derivativesMicrobiological testing/measurementRapid identificationOligonucleotide Primer
The present invention provides oligonucleotide primers and compositions and kits containing the same for rapid identification of bacteria by amplification of a segment of bacterial nucleic acid followed by molecular mass analysis.
Owner:IBIS BIOSCI
Compositions and methods for detecting BV-associated bacterial nucleic acid
ActiveUS9657352B2Opens up structureStrong specificityMicrobiological testing/measurementBacterial vaginosisOligomer
Disclosed are nucleic acid oligomers, including amplification oligomers, capture probes, and detection probes, for detection of a 16S rRNA or its encoding gene from bacterial species associated with bacterial vaginosis. Also disclosed are methods of specific nucleic acid amplification and detection using the disclosed oligomers, as well as corresponding reaction mixtures and kits.
Owner:GEN PROBE INC
Digital loop-mediated isothermal amplification (LAMP) method based on track etching film and application
InactiveCN110846384ASimplify the experimental stepsEasy to operateMicrobiological testing/measurementNanoreactorFluorescence
The invention discloses a digital loop-mediated isothermal amplification (LAMP) method based on track etching film and application and relates to the technical field of molecular biology. The method is characterized in that pores in the track etching film are used as isolated nano reactors, a single bacterium is evenly dispersed into each hole of the film to perform LAMP to generate fluorescence,and ImageJ software is used to perform fluorescence photographing counting on the film system after the isothermal amplification to quantitatively calculate the original nucleic acid concentration ofto-be-measured objects in the film system so as to achieve absolute quantitative analysis of the bacteria. The method has the advantages that bacterium nucleic acid digital analysis can be completed without equipment such as complex micro-fluidic devices and chips which are high in cost, the method is simple in experiment steps, fast and efficient, simple to operate and low in cost, is not dependent on the target bacteria and can carry on the digital detection of the target bacteria only by designing matched primers, and a digital nucleic acid detection technology which is cheap and flexible and simple to operate is provided for gene research.
Owner:ZHEJIANG UNIV
Methods for identifying markers of antimicrobial compounds
The present invention relates to arrays for bacterial gene expression profiling in response to treatment with antimicrobial compounds. Bacterial nucleic acid molecules are used as the hybridizable elements in various applications, such as arrays for gene expression profiling and for determining a mode of action for an antimicrobial compound. The present invention also relates to nucleic acid sequences which can be developed as targets for drug screening.
Owner:NOVO NORDISKBIOTECH INC
Respiratory syncytial virus expression vectors
ActiveUS10227569B2SsRNA viruses negative-senseAnimal cellsNucleic acid sequencingRespiratory syncytial virus (RSV)
In certain embodiments, the disclosure relates to vectors containing bacterial nucleic acid sequences and a paramyxovirus gene. Typically, the expression vector comprises a bacterial artificial chromosome (BAC), and a nucleic acid sequence comprising a respiratory syncytial virus (RSV) gene in operable combination with a regulatory element and optionally a reporter gene.
Owner:EMORY UNIVERSITY +1
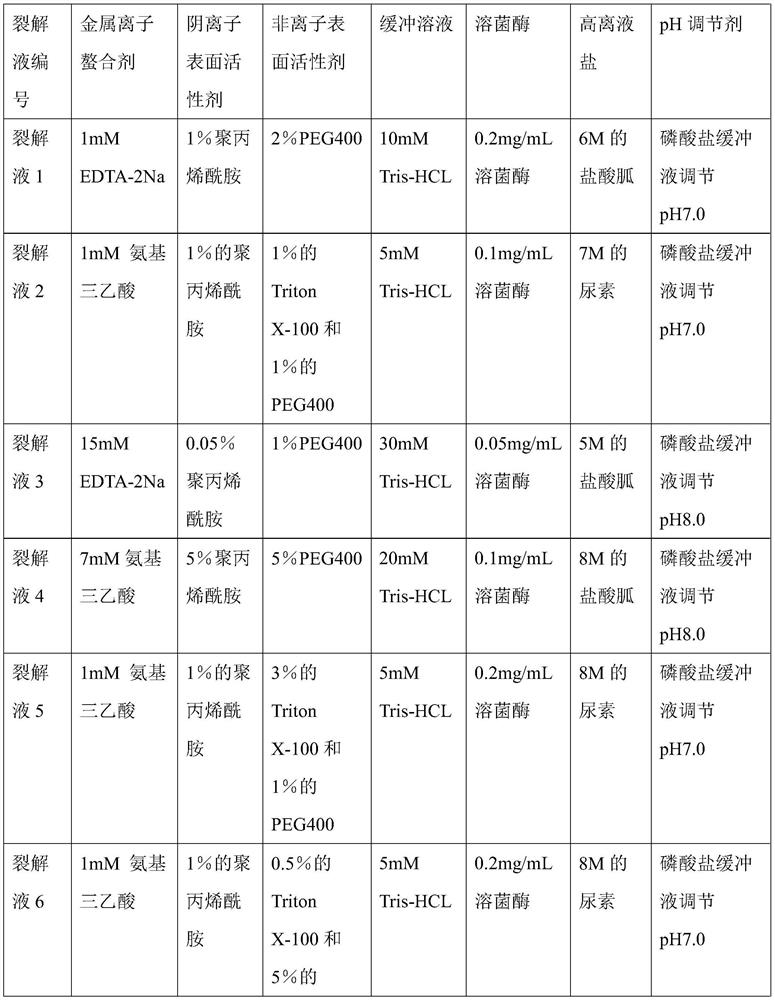
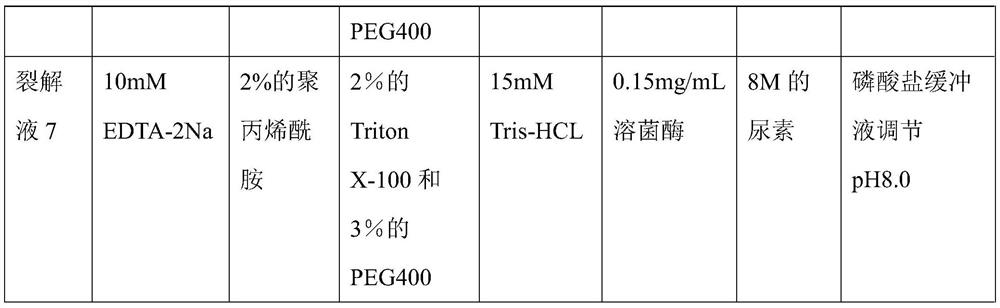
Bacterial nucleic acid extraction lysate as well as preparation method and application thereof
The invention provides a bacterial nucleic acid extraction lysate, which comprises the following components: 1-15 mM of a metal ion chelating agent, 0.05-5% of an anionic surfactant, 0.5-5% of a nonionic surfactant, 5-30 mM of a buffer solution, 0.05-0.2 mg / mL of lysozyme, 5-8 M of a high ionic liquid salt, a pH regulator and a solvent. And the pH regulator is used for regulating the pH of the lysis solution to 7-8. The method is simple, convenient and rapid to operate and low in cost, can provide a basis for nucleic acid amplification detection, provides support for rapid diagnosis of gastrointestinal tract related diseases, also reduces the time cost and economic cost in the definite diagnosis process, has broad market prospects and great economic and social benefits, and is suitable forlarge-scale popularization and application.
Owner:广州源古纪科技有限公司
Kit for rapidly extracting bacterial nucleic acid based on magnetic beads and extraction method
The invention discloses a kit for rapidly extracting the bacterial nucleic acid based on magnetic beads and an extraction method. Reagents of the kit include a lysis solution, a magnetic bead solution, a deproteinization rinsing solution, a desalination rinsing solution and a nucleic acid eluent. The method comprises the steps of adding the magnetic bead solution and the lysis solution into a sample for lysis, performing rinsing with the rinsing solutions, and performing eluting with the nucleic acid eluent to obtain the bacterial nucleic acid. The kit for rapidly extracting bacterial nucleic acid based on the magnetic beads according to the invention is simple and convenient to extract, does not contain toxic reagents such as phenol chloroform, can be manually operated, can be integrated with various automatic nucleic acid extractors, has simple and convenient steps, and can automatically extract the nucleic acid only by adding a sample and without adding protease K, simplifies experimental operation steps to the greatest extent and guarantees the safety of an experimenter. The requirements on the nucleic acid extraction speed and flux and the requirements on the nucleic acid extraction quality when more clinical samples exist in a hospital are met; and meanwhile, the kit according to the invention is low in toxicity, simple and convenient to operate, high in sample flux and better in nucleic acid extraction effect.
Owner:GENFINE BIOTECH BEIJING CO LTD
Compositions and methods for detecting BV-associated bacterial nucleic acid
ActiveUS9663829B2Opens up structureStrong specificitySugar derivativesMicroorganismsBacterial vaginosisOligomer
Disclosed are nucleic acid oligomers, including amplification oligomers, capture probes, and detection probes, for detection of a 16S rRNA or its encoding gene from bacterial species associated with bacterial vaginosis. Also disclosed are methods of specific nucleic acid amplification and detection using the disclosed oligomers, as well as corresponding reaction mixtures and kits.
Owner:GEN PROBE INC
Method and system for determining a bacterial resistance to an antibiotic drug
The invention relates to a method, a databank, a system and a computer program product for determining a bacterial resistance to an antibiotic drug. A data bank is provided which comprises a plurality of bacterial reference nucleic acid sequences, wherein at least some reference nucleic acid sequences are associated with a respective antibiotic drug resistance information. A comparison unit is used for comparing said bacterial nucleic acid sequence information obtained from a sample with said plurality of bacterial reference nucleic acid sequences.
Owner:ARES GENETICS GMBH
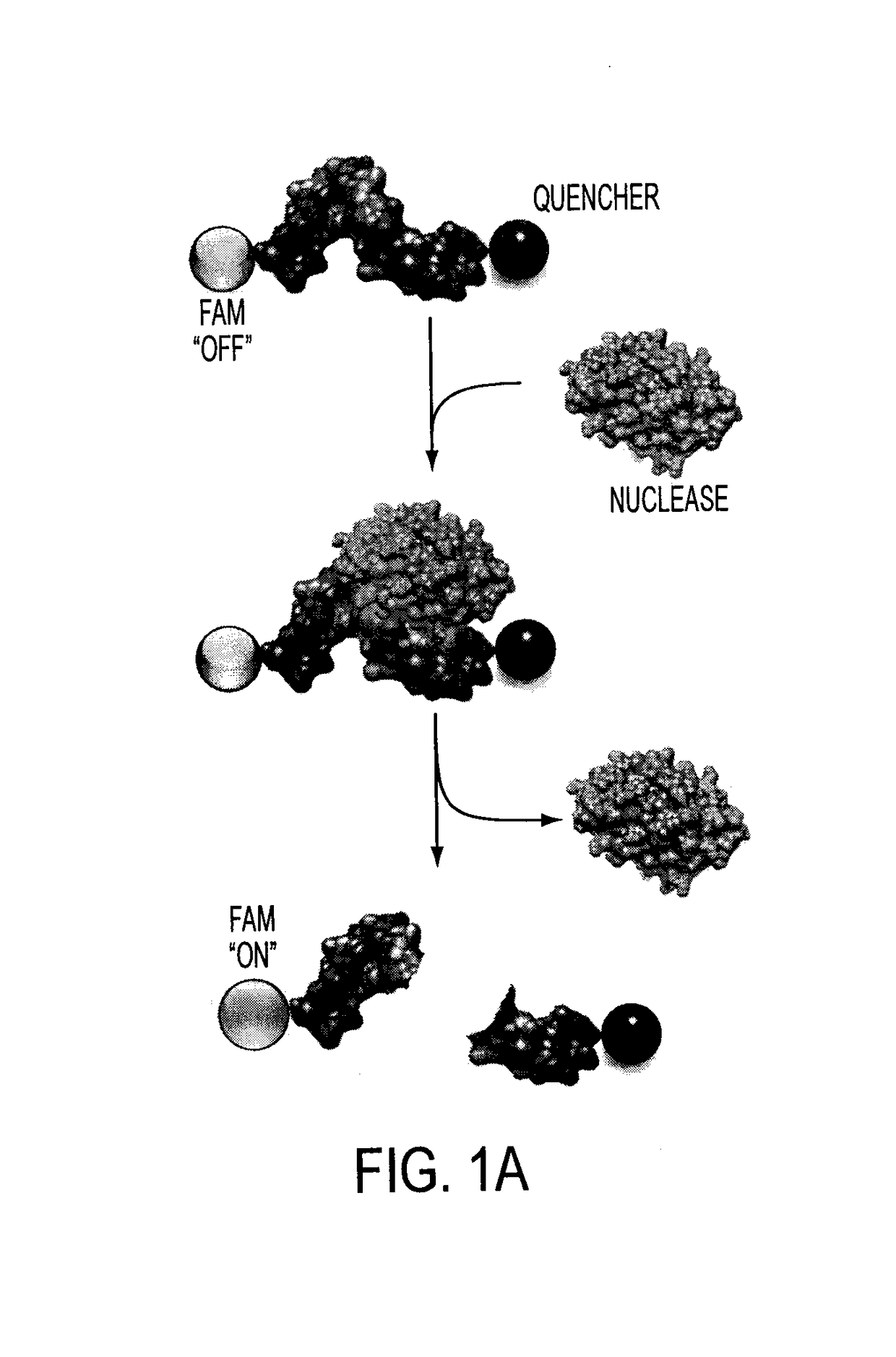
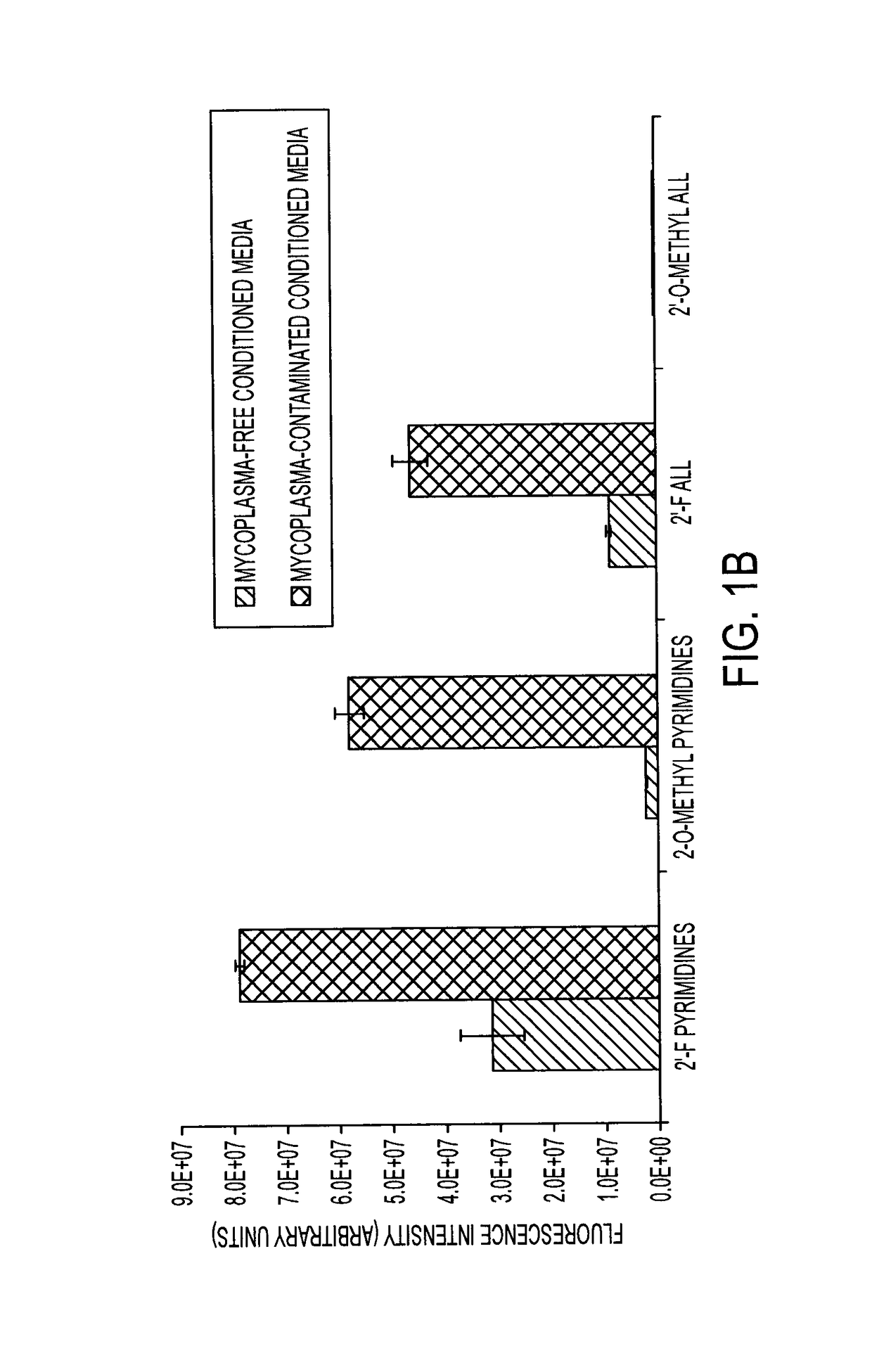
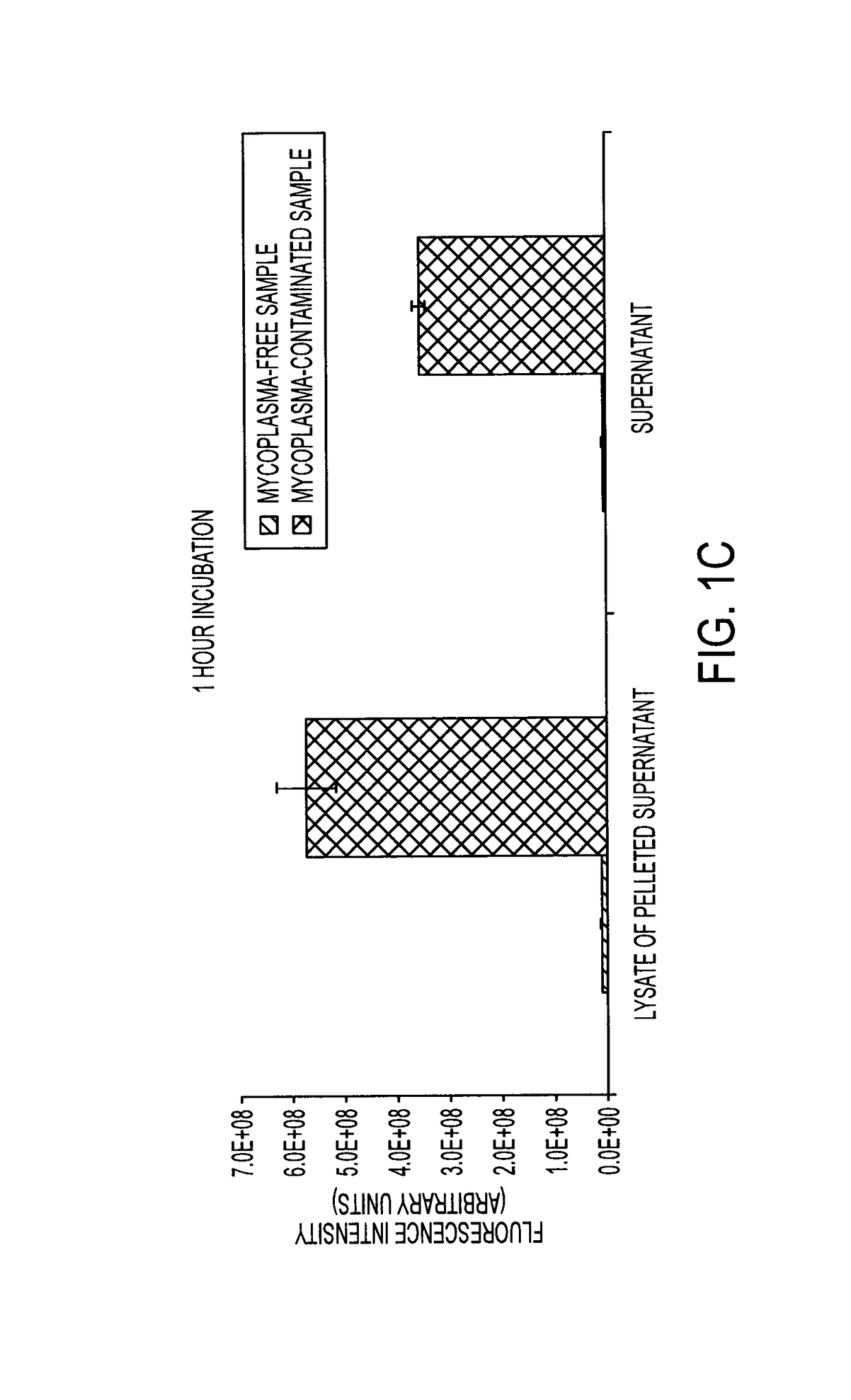
Oligonucleotide-based probes for detection of bacterial nucleases
The present invention relates to a rapid detection of microbial-associated nuclease activity with chemically modified nuclease (e.g., ribonuclease) substrates, and probes and compositions useful in detection assays. Accordingly, in certain embodiments, the present invention provides a probe for detecting a microbial endonuclease comprising a substrate oligonucleotide of 2-30 nucleotides in length, a fluorescence-reporter group operably linked to the oligonucleotide, and a fluorescence-quencher group operably linked to the oligonucleotide. The fluorescence-reporter group and the fluorescence-quencher group are separated by at least one RNAse-cleavable residue, e.g., RNA base.
Owner:UNIV OF IOWA RES FOUND +1
Process for abstracting bacterial DNA from phlegm, kit and uses thereof
ActiveCN101285062BEasy to operateReduce the amount of solutionMicrobiological testing/measurementDNA preparationWild typeSolid particle
The invention discloses a method and a kit for extracting bacterial nucleic acid from sputa and applications thereof. The method is to use liquefaction reagent to liquefy a sputum sample and add solid particles to lyse cells to extract nucleic acid form the sputum. Overcoming the drawbacks of the prior method including complex procedures, a long period of time, complex operation and inadequate samples, the method of the invention for extracting nucleic acid samples is convenient and quick in operation, adequate in nucleic acid samples, low in cost and easy to implement. The method and kit forextracting bacterial nucleic acid can be used to identify strains of mycobacteria and to detect whether a gene locus of mycobacterium tuberculosis is of a wide type or a mutant type. With a chip for identifying strains and a chip for detecting whether the gene locus of mycobacterium tuberculosis is of a wide type or a mutant type, the method can identify the strains of mycobacteria and to detect whether a gene locus of mycobacterium tuberculosis is of a wide type or a mutant type in a quick, accurate and high-pass way.
Owner:CAPITALBIO CORP
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com