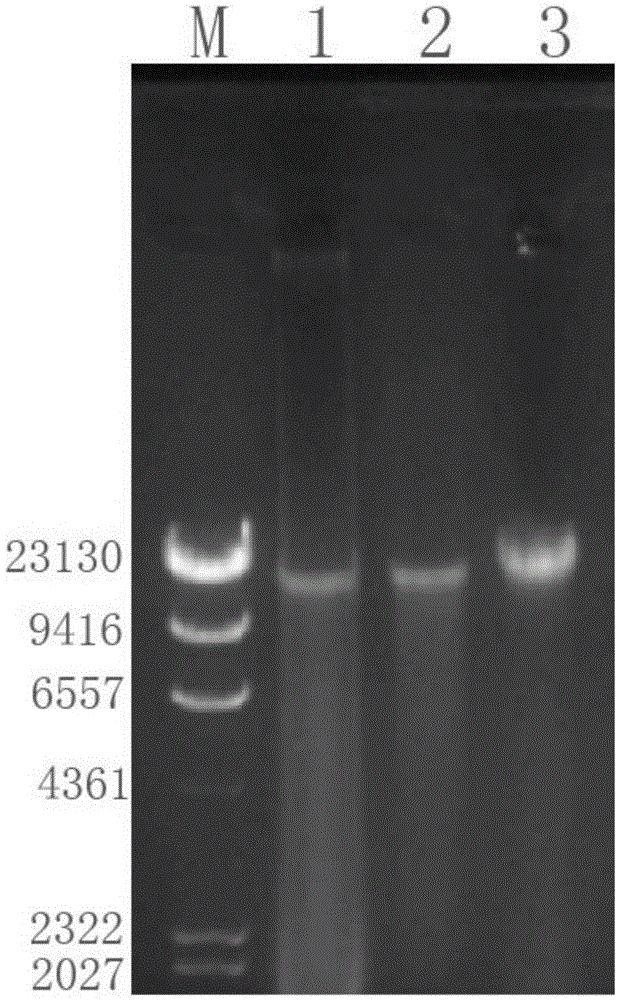
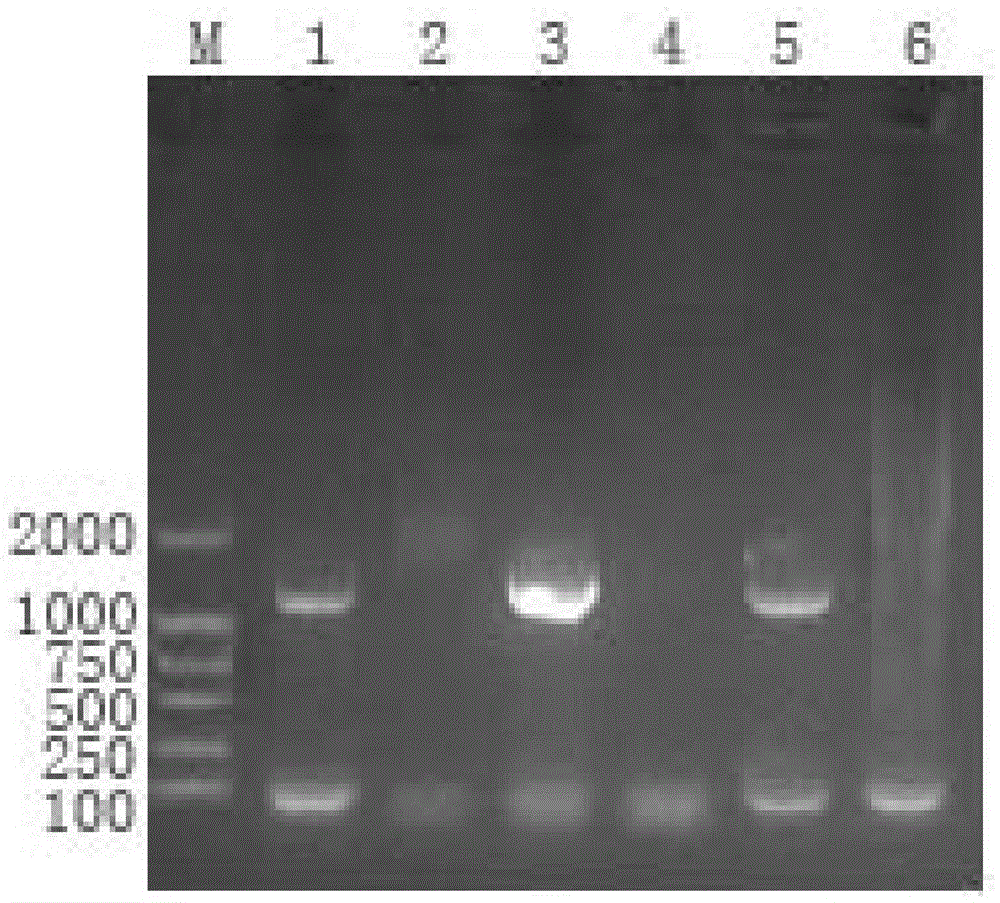
Bacteriophage genome DNA (deoxyribonucleic acid) extraction kit and method
An extraction method and phage technology, which is applied in the field of bacteriophage genomic DNA extraction kits, can solve the problems of reducing the operating time of technicians, and achieve the effects of rapid extraction, high extraction yield, and eradication of interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Extracting the Genomic DNA of Vibrio alginolyticus Phage ФPE333
[0041] Prepare reagents according to the following formula:
[0042] (1) Reagent A: chloroform;
[0043] (2) Reagent B: 20mg / ml DNaseI prepared with TE buffer, stored at -20°C;
[0044] (3) Reagent C: 20mg / ml RNaseA prepared with TE buffer, stored at -20°C;
[0045] (4) Reagent D: Precipitation buffer containing 400 mg / ml polyethylene glycol-8000 (PEG-8000) and 3M NaCl, stored at room temperature;
[0046] (5) Reagent E: lysis buffer, weigh 5.8gNaCl, 1.23gMgSO 4 ·7H 2 O, prepare 50ml of 1MTris-Cl (1MTris, pH adjustment 8.0), 100ml of 0.5MEDTA, mix the above components and add ddH 2 0 to 1000ml, autoclave for 20min;
[0047] (6) Reagent F: as the main lysate, 200mg / ml sodium dodecylbenzenesulfonate (SDS);
[0048] (7) Reagent G: an auxiliary lysate, 25 mg / ml proteinase K prepared with TE buffer, stored at 4°C;
[0049] (8) Reagent H: impurity removal solution, 6MNaCl;
[0050] (9) Reage...
Embodiment 2
[0064] Example 2: Extraction of coliphage ФP1655 genomic DNA
[0065] Prepare reagents according to the following formula:
[0066] (1) Reagent A: chloroform;
[0067] (2) Reagent B: 10mg / ml DNaseI prepared with TE buffer, stored at -20°C;
[0068] (3) Reagent C: 10mg / ml RNaseA prepared with TE buffer, stored at -20°C;
[0069] (4) Reagent D: Precipitation buffer containing 300 mg / ml polyethylene glycol-8000 (PEG-8000) and 3M NaCl, stored at room temperature;
[0070] (5) Reagent E: lysis buffer, weigh 5.8gNaCl, 1.23gMgSO 4 ·7H 2 O, prepare 50ml of 1MTris-Cl (1MTris, pH adjustment 8.0), 100ml of 0.5MEDTA, mix the above components and add ddH 2 0 to 1000ml, autoclave for 20min;
[0071] (6) Reagent F: as the main lysate, 150mg / ml sodium dodecylbenzenesulfonate (SDS);
[0072] (7) Reagent G: an auxiliary lysate, 15 mg / ml proteinase K prepared with TE buffer, stored at 4°C;
[0073] (8) Reagent H: impurity removal solution, 5M NaCl;
[0074] (9) Reagent I: DNA binding bu...
Embodiment 3
[0088] Example 3: Extracting Genomic DNA of Enterobacter cloacae Phage ФPZJ02
[0089] Prepare reagents according to the following formula:
[0090] (1) Reagent A: chloroform;
[0091] (2) Reagent B: 15mg / ml DNaseI prepared with TE buffer, stored at -20°C;
[0092] (3) Reagent C: 15mg / ml RNaseA prepared with TE buffer, stored at -20°C;
[0093](4) Reagent D: Precipitation buffer containing 330 mg / ml polyethylene glycol-8000 (PEG-8000) and 3M NaCl, stored at room temperature;
[0094] (5) Reagent E: lysis buffer, weigh 5.8gNaCl, 1.23gMgSO 4 ·7H 2 O, prepare 50ml of 1MTris-Cl (1MTris, pH adjustment 8.0), 100ml of 0.5MEDTA, mix the above components and add ddH 2 0 to 1000ml, autoclave for 20min;
[0095] (6) Reagent F: as the main lysate, 180mg / ml sodium dodecylbenzenesulfonate (SDS);
[0096] (7) Reagent G: an auxiliary lysate, 20 mg / ml proteinase K prepared with TE buffer, stored at 4°C;
[0097] (8) Reagent H: impurity removal solution, 5.5M NaCl;
[0098] (9) Reagent...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com