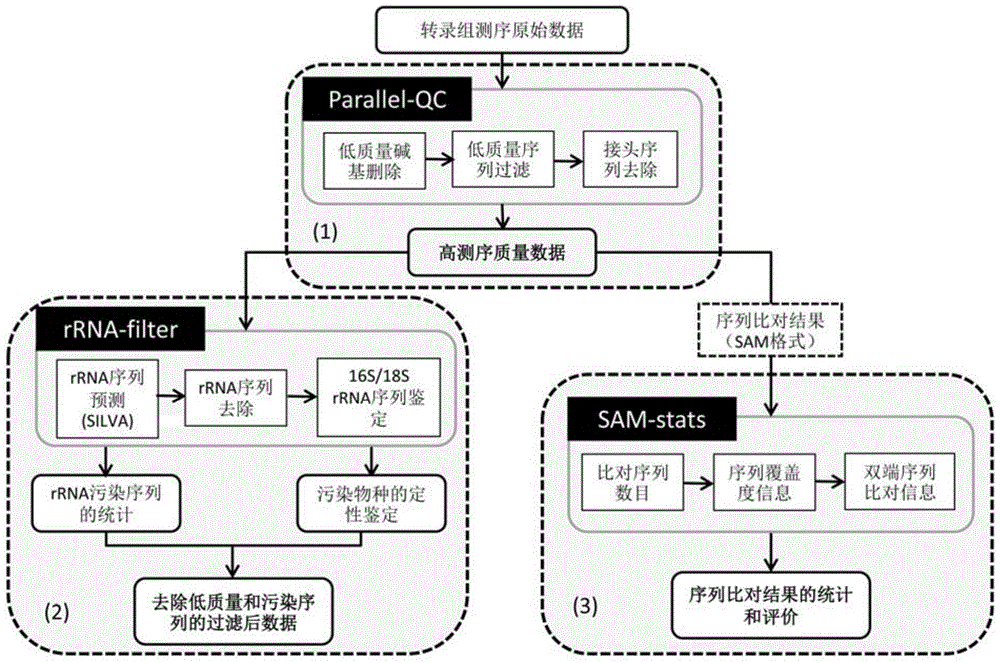
High-flux transcriptome sequencing data quality control method based on multi-core CPU (Central Processing Unit) hardware
A transcriptome sequencing and data quality technology, applied in the field of bioinformatics, can solve the problem that the computing system cannot efficiently meet the quality control of high-throughput transcriptome sequencing data, so as to overcome the bottleneck of computing efficiency, improve efficiency, and improve accuracy and speed effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0034] The present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments.
[0035] The technical scheme adopted by the invention is a multi-core CPU computer and an efficient and unified software platform built on it. Its characteristics are (1) a high-performance parallel computing and storage hardware system; (2) a comprehensive, high-performance, unified, and configurable parallel software platform.
[0036] (1) High-performance parallel computing and storage hardware
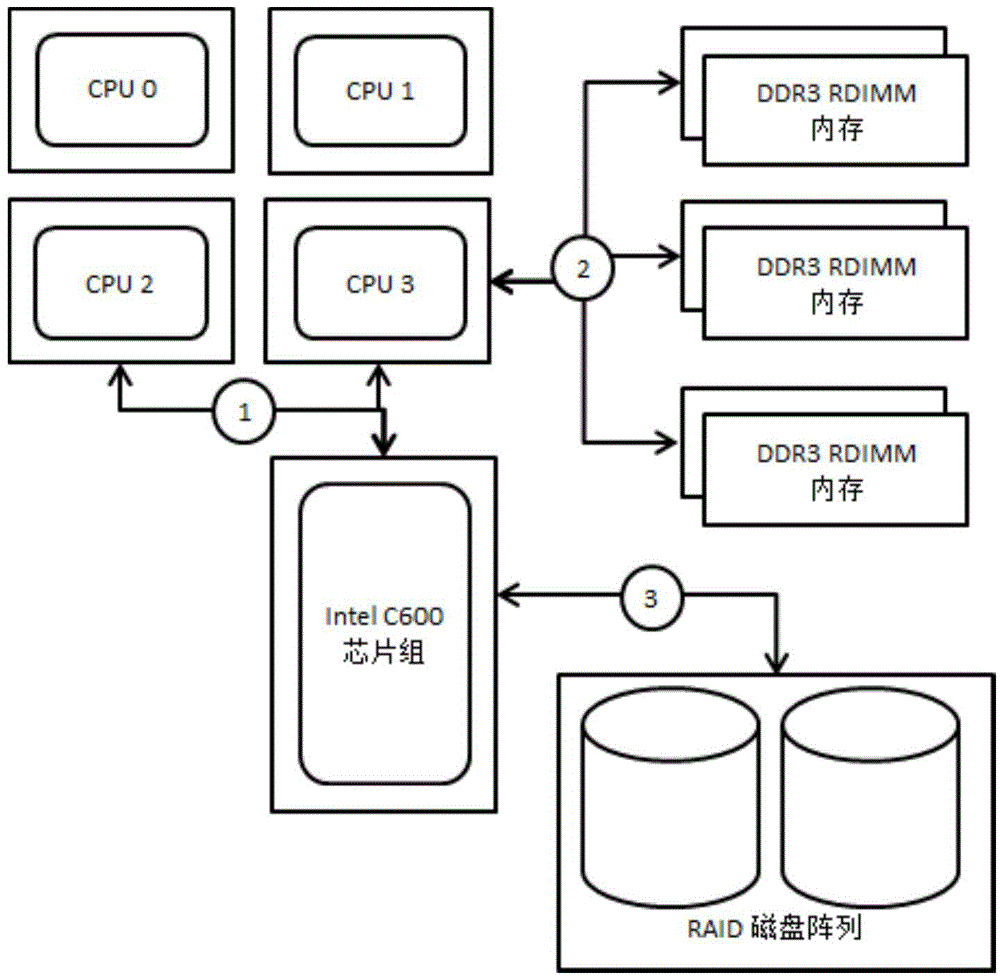
[0037] The hardware system uses multi-channel multi-core CPU for large-scale parallel computing. figure 1 It is the system structure diagram of the computing server:
[0038] First of all, multi-channel multi-core CPU parallelizes calculations, using 4-way processors, and using QPI bus connections between processors. Each processor has 8 independent computing cores, equipped with three-channel DDR3RDIMM memory, and also adapts to the computing re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com