Genome assembling method
A genome assembly and consistency technology, applied in the field of genome assembly, can solve problems such as indistinguishability, fragmentation of continuous sequence fragments, and many operation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0056] The present invention will be further described below in conjunction with the accompanying drawings and specific embodiments.
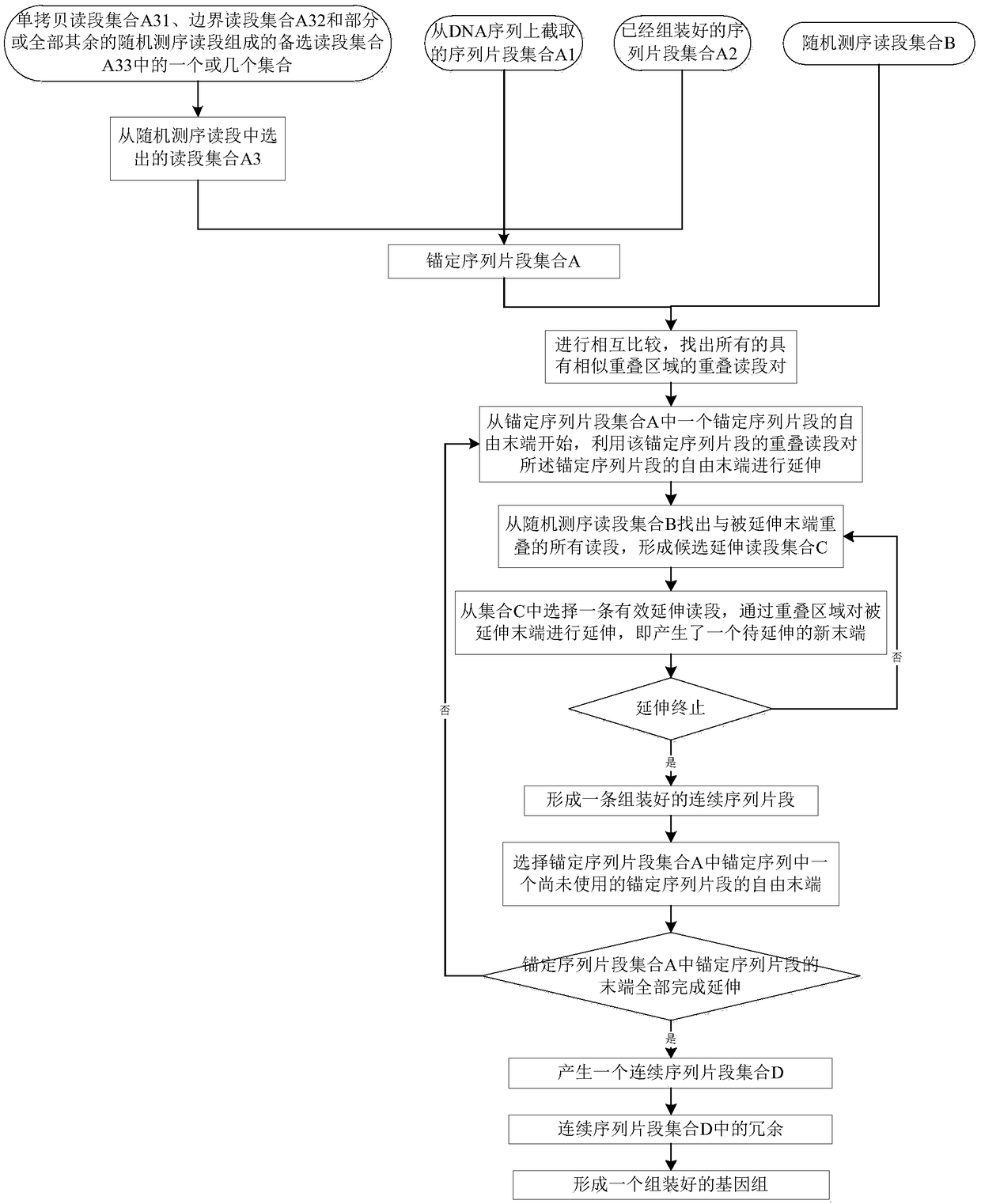
[0057] Embodiments of the present invention, a genome assembly method, such as figure 1 shown, including the following steps:
[0058] S1, compare all known DNA sequence fragments with each other, and find all overlapping read pairs with similar overlapping regions; wherein, the known DNA sequences include anchor sequence fragment set A and random sequencing read set B; the set of anchor sequence fragments A includes: one of the sequence fragment set A1 intercepted from the DNA sequence, the sequence fragment set A2 that has been assembled, and the read segment set A3 selected from random sequencing reads, or Several collections; said comparing all known DNA sequence fragments to each other, including comparing all anchor sequence fragments to all sequencing reads to each other, comparing all sequencing reads to each other, or comparing all T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com