Optimization method for accurate species identification of human flora through high-throughput 16S rDNA sequencing
An optimization method and high-throughput technology, applied in the fields of medicine and molecular biology, can solve problems such as lack of relative abundance information and inability to form sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
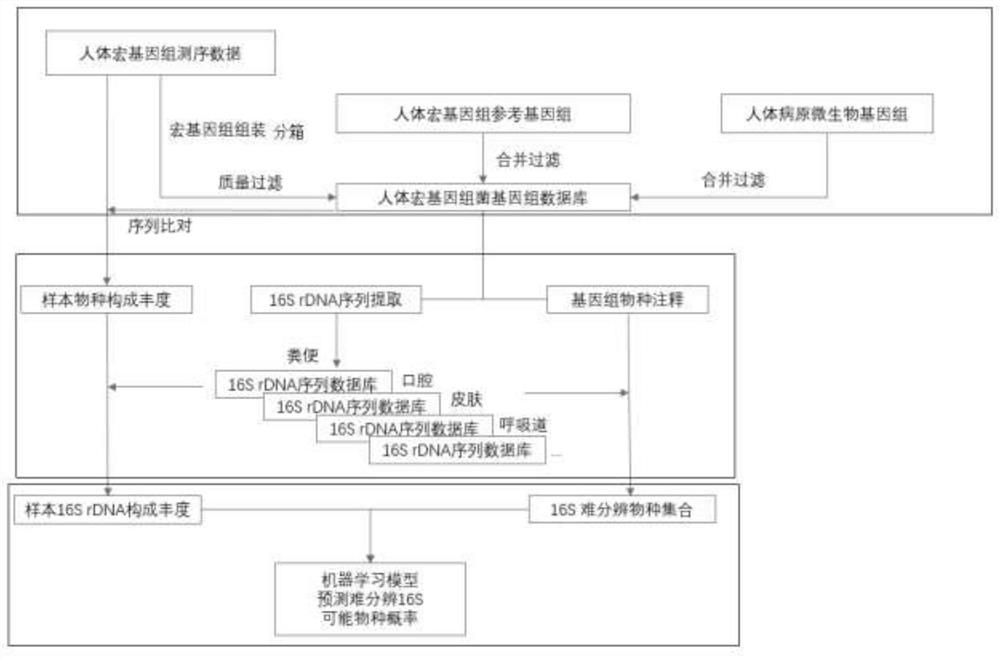
[0076] Example 1. An optimized method for the accurate identification of human flora 16S rDNA high-throughput sequencing species, such as figure 1 described, including the following steps:
[0077] step 1),
[0078] 1.1) According to the WGS method, the samples from the same human tissue source are sequenced, so as to establish a human metagenomic sequencing database based on human microbial samples, so the human metagenomic sequencing data meet the following conditions: the species is human, and the sequencing method is WGS ( Whole genome sequencing), with a clear source of samples.
[0079] Sources of human metagenomic sequencing data that meet the above requirements include but are not limited to:
[0080] NCBI's sra database https: / / www.ncbi.nlm.nih.gov / sra;
[0081] https: / / www.ebi.ac.uk / metagenomics / ;
[0082] Explanation: The above two databases are existing metagenomic open data storage databases, which can be directly downloaded from the above to obtain the public...
Embodiment 2
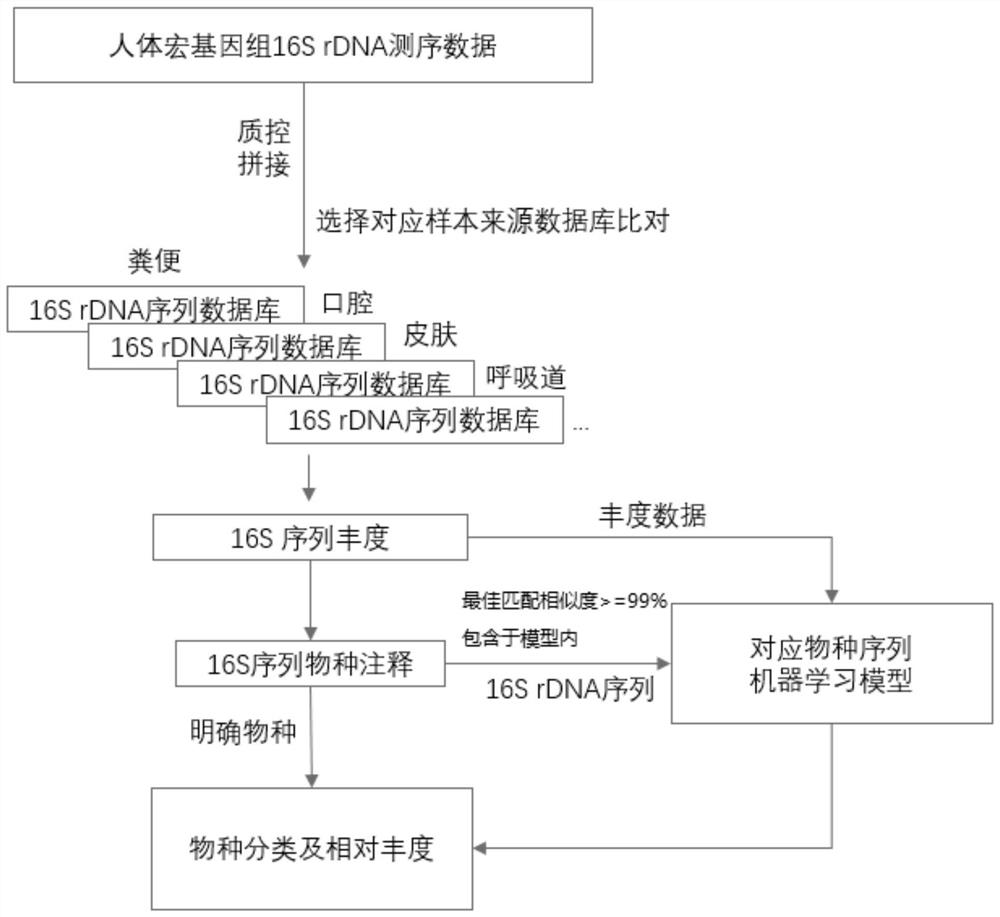
[0107] Embodiment 2, this is the use of species identification and abundance calculation of high-throughput sequencing 16S rDNA sequences based on the database and model of the above-mentioned embodiment 1; specific operations include the following steps:
[0108] 1) Obtain sample high-throughput sequencing 16S rDNA sequence: Obtain the full-length or partial segment sequencing sequence of 16S rDNA through second-generation or third-generation high-throughput sequencing, and then follow the quality inspection and splicing of the above steps to obtain human 16S rDNA sequencing data .
[0109] The sample has a clear source of the human tissue part of the sample.
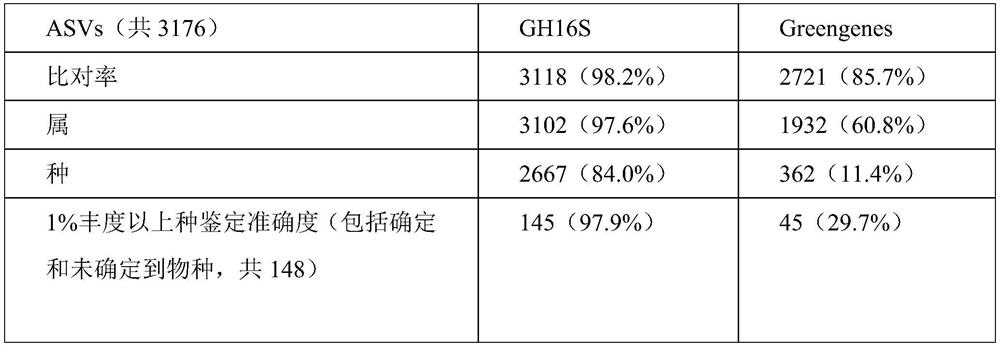
[0110] 2), first perform sequence alignment according to the type of sample source and the corresponding 16S species sequence database (16srDNA sequence database) in step 2) of Example 1 (common 16S comparison and analysis software such as DADA2 or Vsearch can be used), so as to obtain 16S sequence abundance degree; a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com