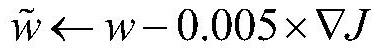Patents
Literature
64 results about "Species classification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Classification of species. In traditional classification, or phylogenetics, species is the taxon base of systematic, whose rank is just below the type. In scientific classification, a species living or having lived is designated following the rules of binomial nomenclature, established by Carl von Linne in the eighteenth century.
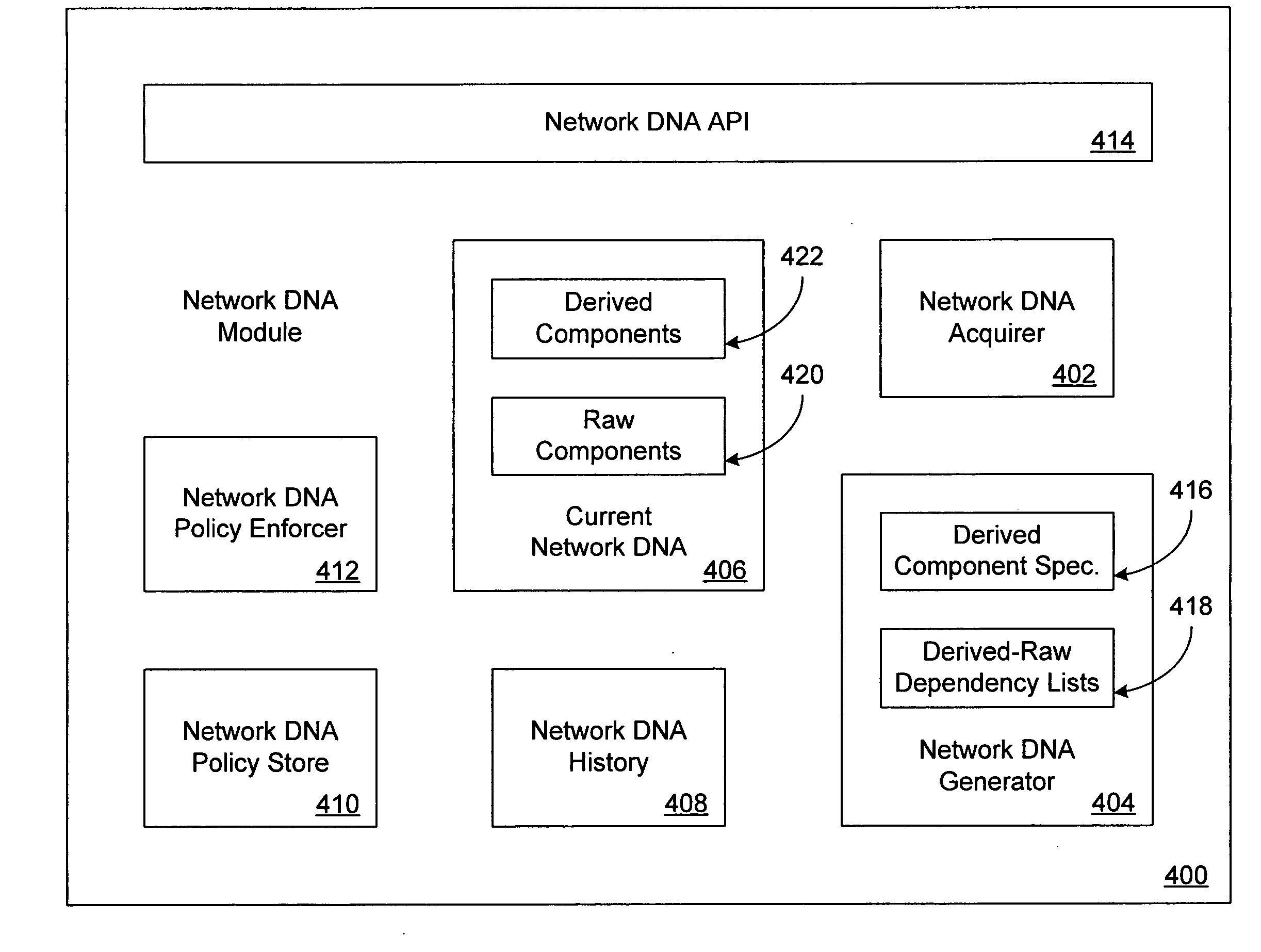
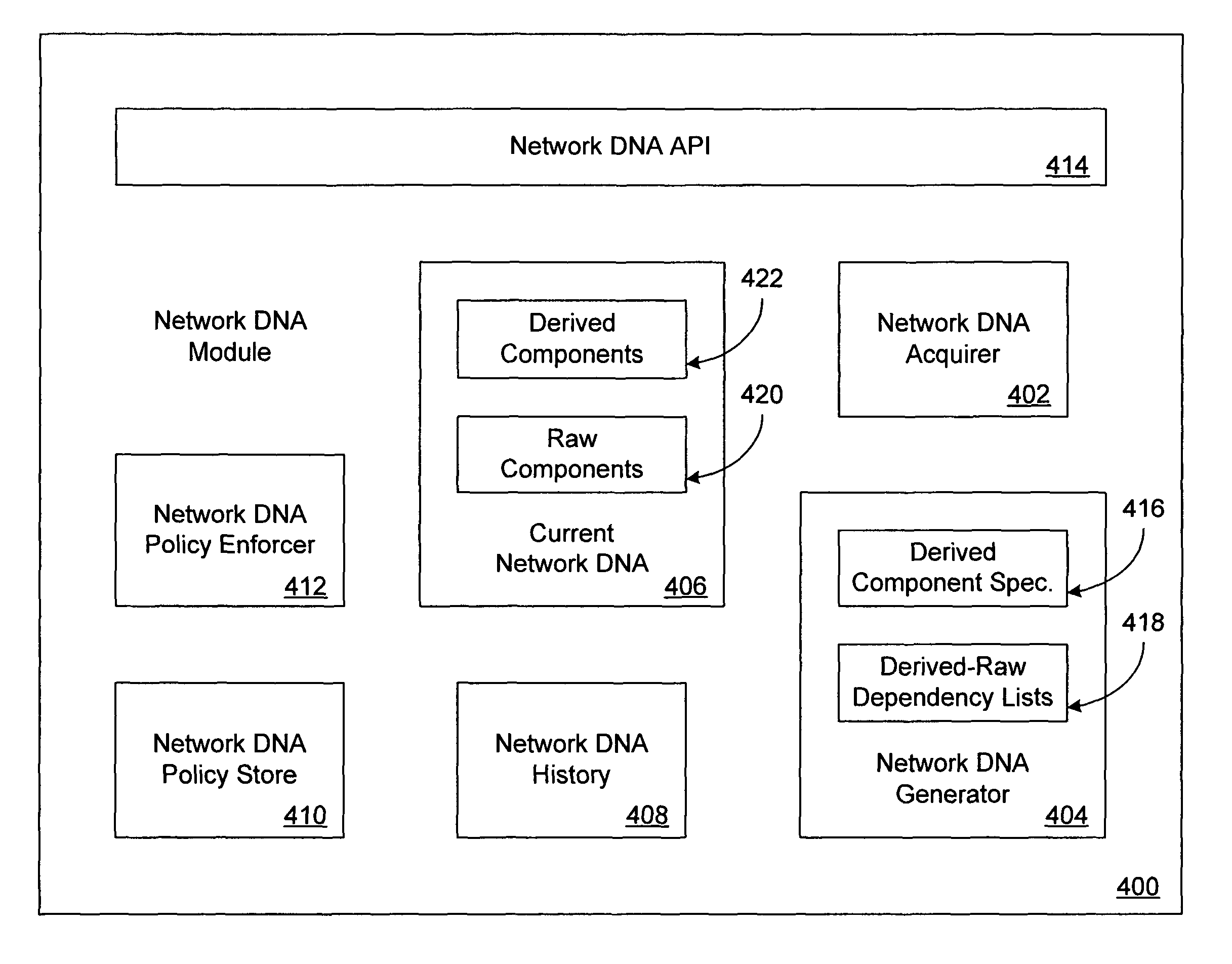
Network DNA
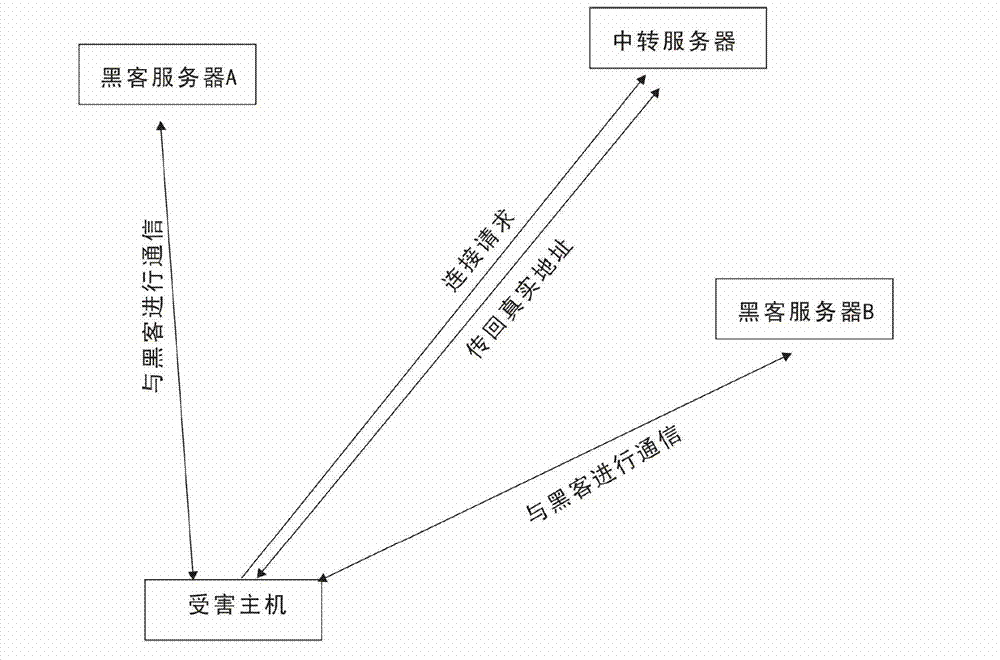
ActiveUS20050177631A1Multiple digital computer combinationsData switching networksPublic placeSpecies classification
Network DNA may be determined for a computer network that taxonomically classifies the computer network. Network DNA may include derived network DNA components and raw network DNA components. Raw network DNA components may be acquired from local or remote sources. Derived network DNA components may be generated according to derived network DNA component specifications. Derived network DNA component specifications may reference raw network DNA components. Network DNA determined for the computer network may include a network species component capable of indicating network species classifications for computer networks. Network species classifications may include enterprise network, home network and public place network. Network species classifications may be determined as a function of network security, network management and network addressing. One or more network DNA stores may be configured to store network DNA for computer networks. Network DNA stores may store network DNA history as well as current network DNA.
Owner:MICROSOFT TECH LICENSING LLC
Currency sorter
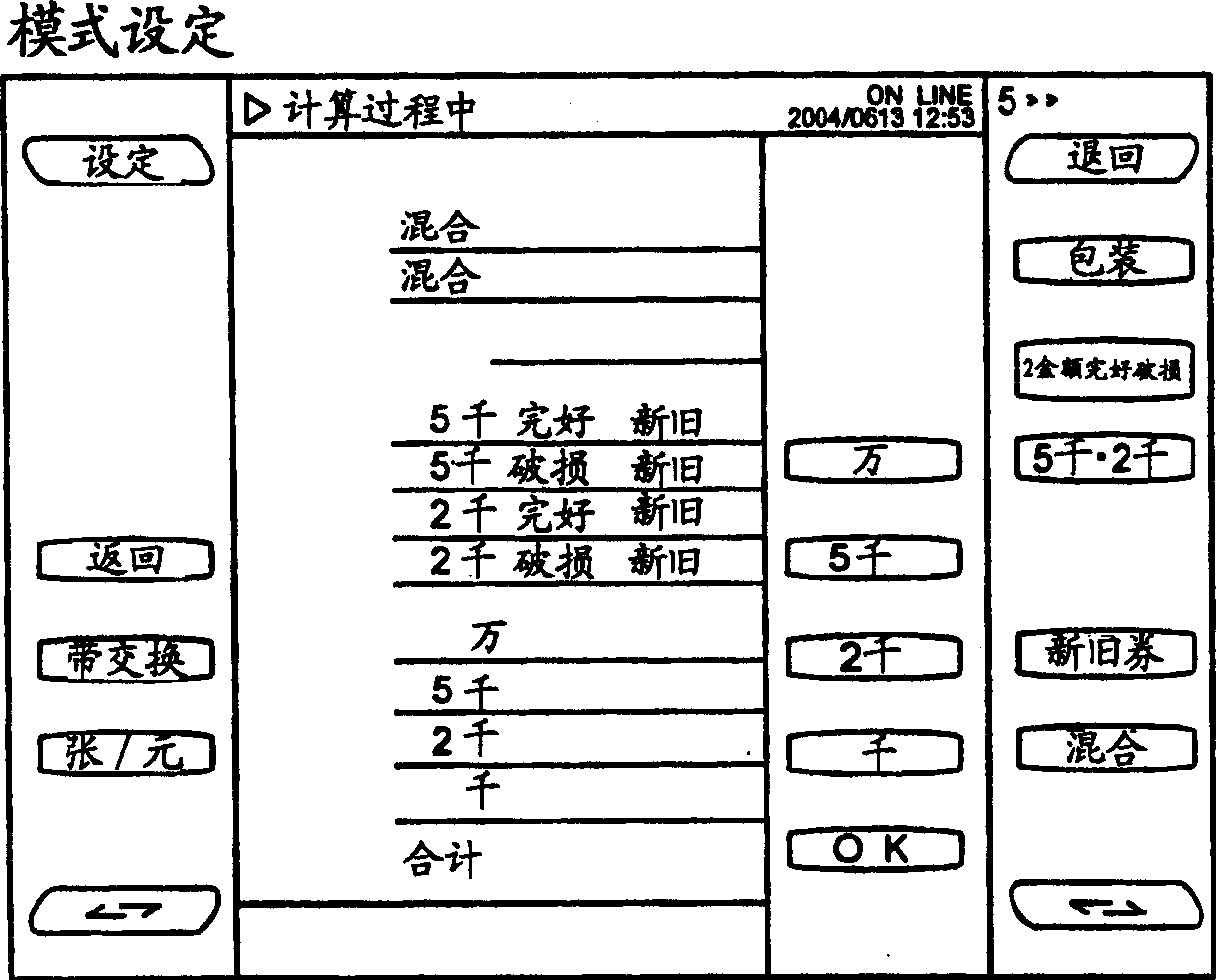
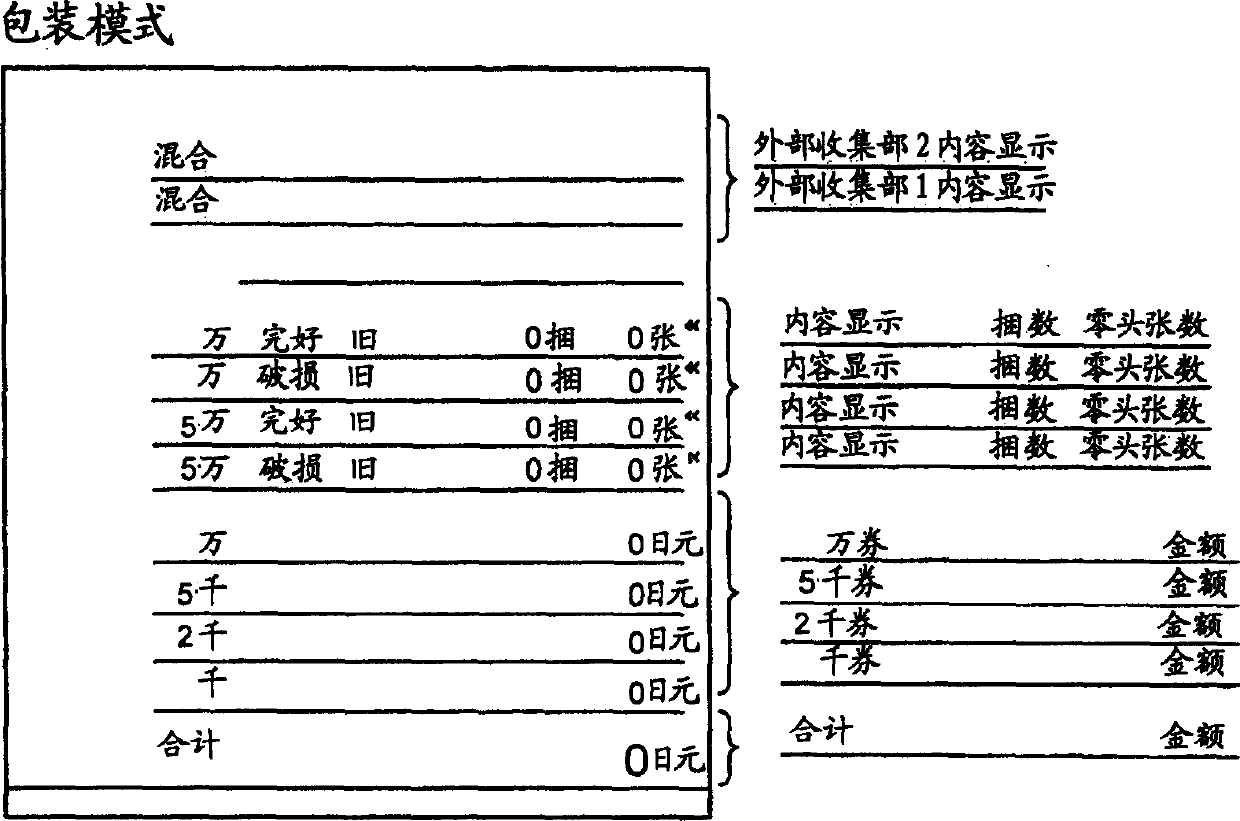
InactiveCN1790426AEasy to handleEffective classificationPaper article packagingCoin/paper handlersSpecies classificationClassification methods
A banknote collating machine has a receiving device for receiving banknotes, a recognition device for recognizing newness, oldness, soundness and damage, and value of the received banknotes, and a plurality of collection parts for collecting predetermined types of banknotes to be bundled based on the recognition results, and specifies the banknotes to be bundled A device for specifying a sorting method for banknotes not to be bundled, and a control unit for distributing the recognized banknotes to the collecting unit and the external collecting unit based on the designation. Preferably there are at least 2 external collection units for collecting banknotes of a specified type that are not subject to binding; the number of collection units is preferably more than the type of banknotes to be bound; it is preferred that the control unit allocates identified banknotes to a plurality of collection units based on the designation of the specifying device. , during the period when one of the multiple collecting parts is full and banknotes of the same amount are collected in the preparatory collecting part, when the other collecting parts are close to being full, the receiving device is switched to intermittent receiving, and the recognition device Confirm one by one until a new reserve collection appears.
Owner:GLORY KOGYO KK
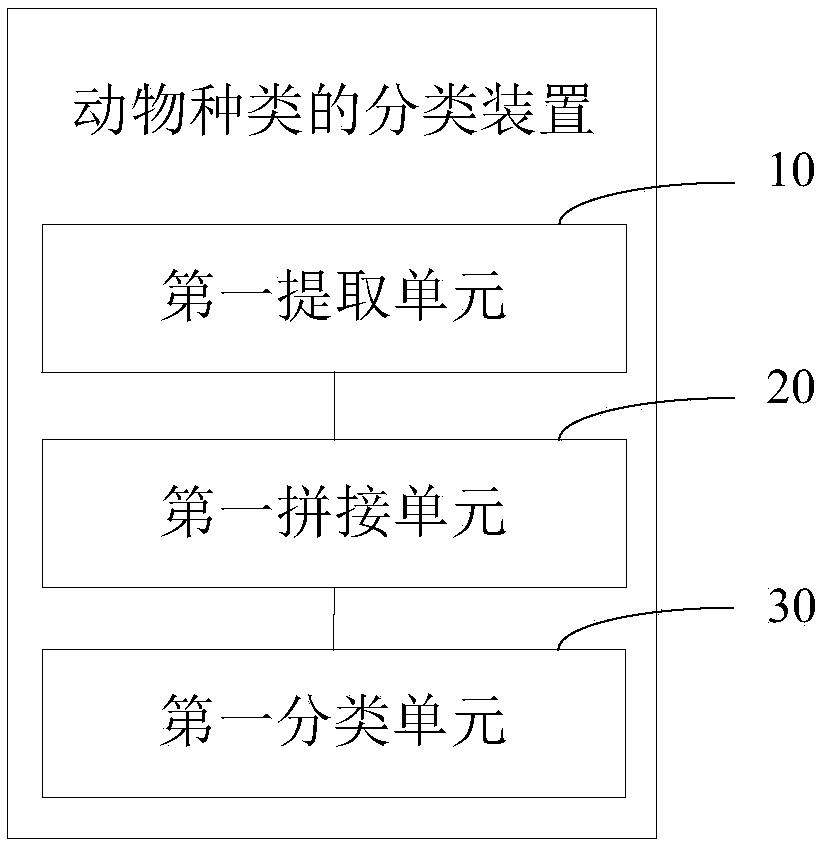
Animal species classification method and device, computer equipment and storage medium
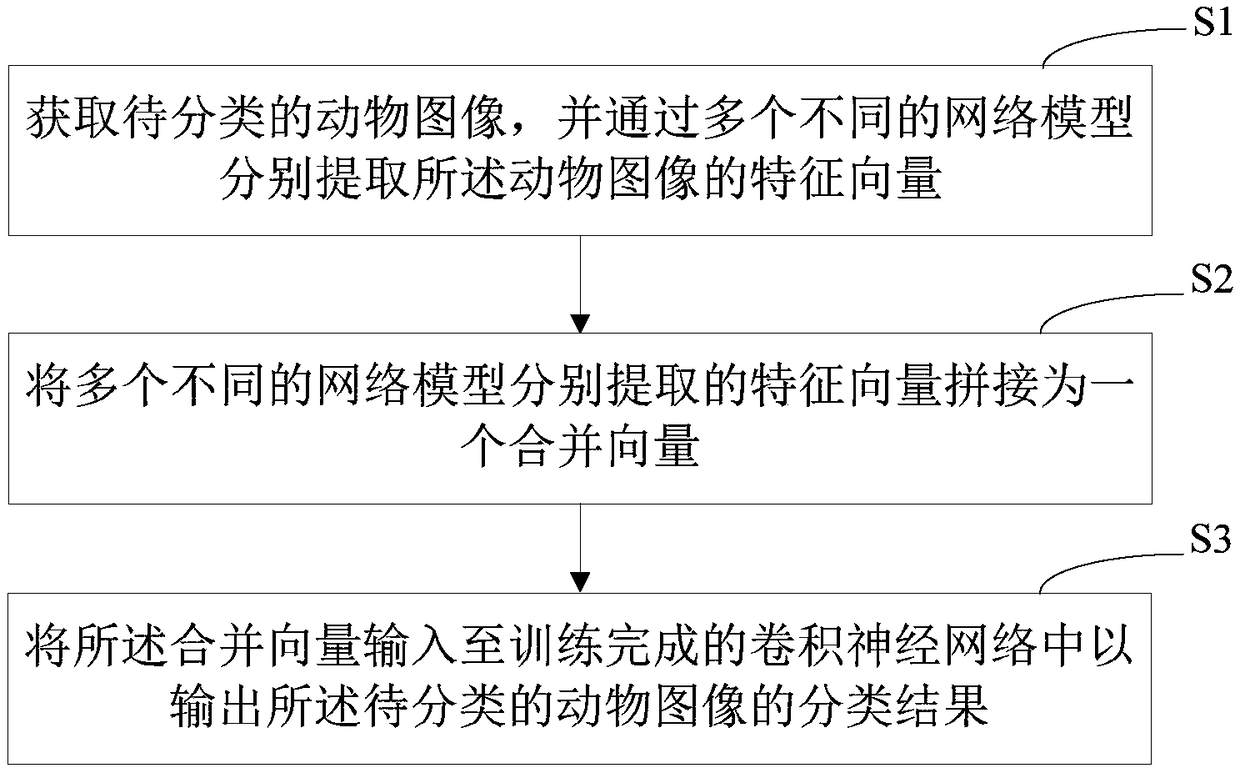
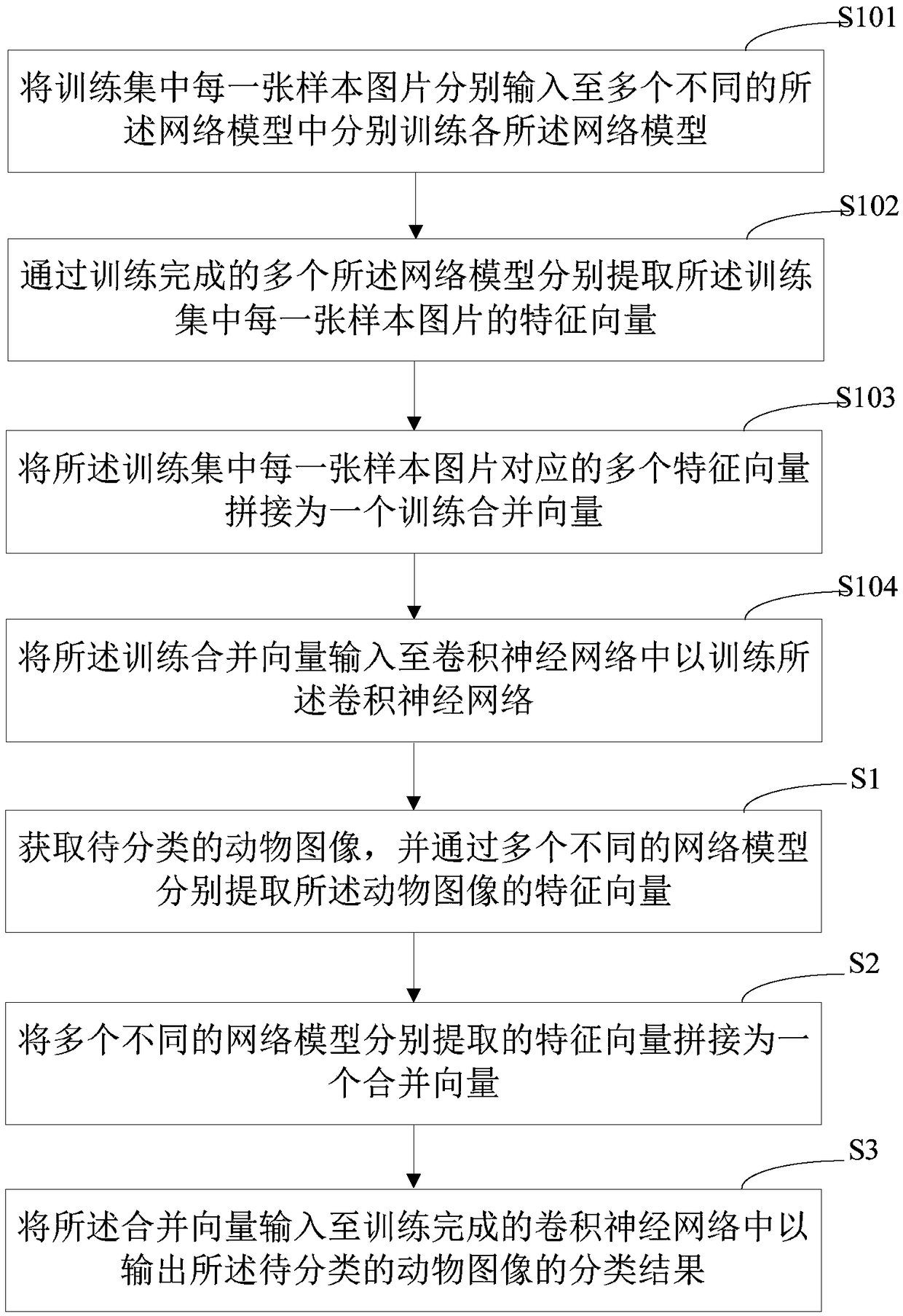
PendingCN108875811AImprove accuracyOvercome the defect of low recognition accuracyCharacter and pattern recognitionNeural architecturesFeature vectorClassification methods
The invention provides an animal species classification method and device, computer equipment and a storage medium. The method comprises the following steps of: obtaining a to-be-classified animal image and respectively extracting feature vectors of the animal image through multiple different network models; splicing the feature vectors respectively extracted by the multiple different network models into a combined vector; and inputting the combined vector into a trained convolutional neural network so as to output a classification result of the to-be-classified animal image. According to theanimal species classification method and device, the computer equipment and the storage medium, the correctness of recognizing animal species is enhanced and the defect that the present recognition correctness is low is overcome.
Owner:PING AN TECH (SHENZHEN) CO LTD
Method for screening veneridae mitochondria COI gene amplification primers
InactiveCN101979536AHigh variabilityLow amplification efficiencyDNA preparationDNA/RNA fragmentationPhylogenesisCoi gene
The invention discloses a method for screening veneridae mitochondria COI (cytochrome c oxidase subunit I) gene amplification primers. The method is characterized by comprising the following steps of: a, logging in National Center for Biotechnology Information, and downloading complete mitochondria sequences of veneridae shellfishes; b, comparing and analyzing the downloaded sequences by using BioEdit, and determining a conserved region of the sequences; c, designing a plurality of pairs of primers by using Primer Premier 5 in the conserved region and synthesizing the primers; d, extracting veneridae shellfish DNA, and performing PCR amplification on the synthesized primers under a corresponding reaction system; and 3, performing electrophoresis detection, and screening a group of cocktail primers with single stripe and high amplification efficiency, wherein the group of cocktail primers consists of a pair of degenerate primers and a pair of common primers. The veneridae mitochondria COI primers can effectively amplify destination fragments of different species of the veneridae, and have significance for COI sequence-based veneridae species classification, phylogenesis, systematicgeography research, veneridae germ plasm resource protection and fishery resource management.
Owner:OCEAN UNIV OF CHINA
Network DNA
ActiveUS8126999B2Multiple digital computer combinationsData switching networksPublic placeNetwork addressing
Network DNA may be determined for a computer network that taxonomically classifies the computer network. Network DNA may include derived network DNA components and raw network DNA components. Raw network DNA components may be acquired from local or remote sources. Derived network DNA components may be generated according to derived network DNA component specifications. Derived network DNA component specifications may reference raw network DNA components. Network DNA determined for the computer network may include a network species component capable of indicating network species classifications for computer networks. Network species classifications may include enterprise network, home network and public place network. Network species classifications may be determined as a function of network security, network management and network addressing. One or more network DNA stores may be configured to store network DNA for computer networks. Network DNA stores may store network DNA history as well as current network DNA.
Owner:MICROSOFT TECH LICENSING LLC
High-throughput mulberry pathogenic bacteria identification and species classification method and application thereof
ActiveCN106636433AMicrobiological testing/measurementMicroorganism based processesDiseaseRibosomal DNA
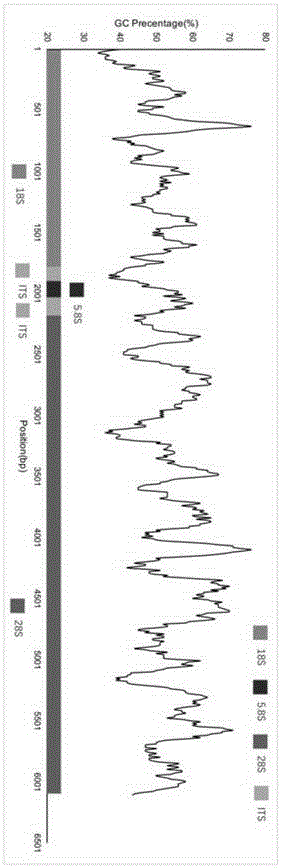
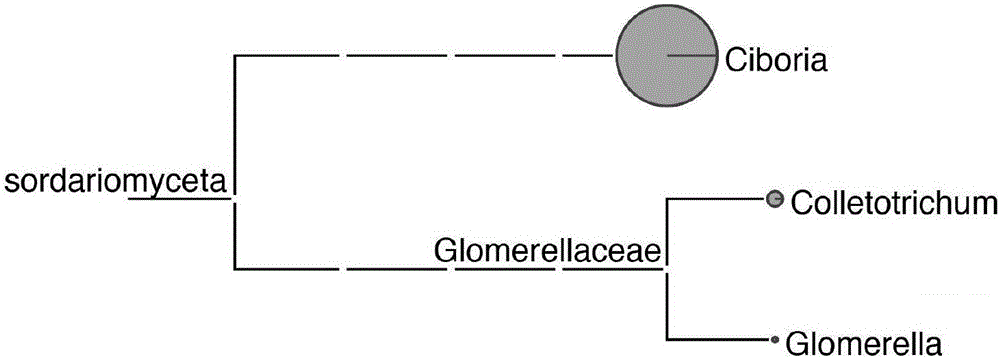
The invention discloses a high-throughput mulberry pathogenic bacteria identification and species classification method. The method comprises the following steps that diseased mulberries are collected; the total DNA of the diseased mulberries is extracted; an Illumina DNA library is created; Illumina high-throughput sequencing is carried out; a mulberry genome sequence in sequencing data is removed; microbial genome sequences are assembled; complete ribosomal DNA sequences are assembled; microbial ribosomal DNA sequences are screened and labeled; the ribosomal DNA sequences are comparatively analyzed to classify species, and thereby the mulberry pathogenic bacteria identification and species classification are fulfilled. A result shows that three species of fungi are identified in total when the method disclosed by the invention is applied to carry out the identification of pathogenic bacteria of popcorn disease and species classification, wherein the Ciboria pathogenic bacteria has the highest relative abundance, hereby the pathogenic Ciboria shiraiana is determined as Ciboria, and according to a comparison result, the pathogenic Ciboria shiraiana is determined as Ciboria carunculoides. Most of the species are phytopathogenic bacteria, and can lead to symptoms, such as mummification and swelling, appearing on fruits and seeds of plants, which are identical with the symptoms of the popcorn disease.
Owner:SOUTH CHINA AGRI UNIV
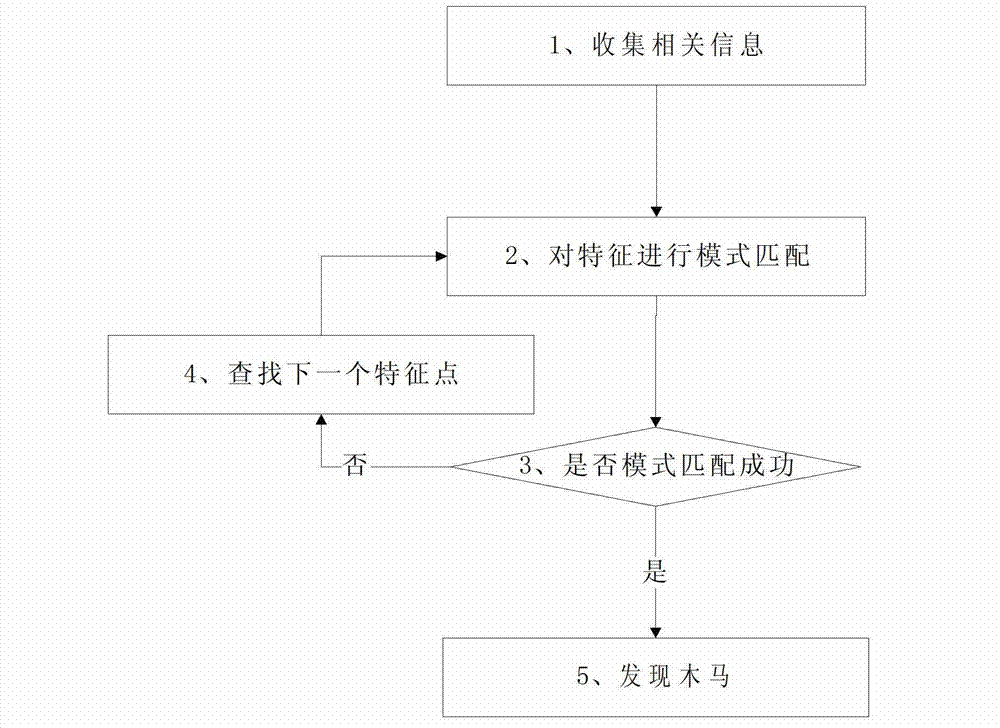
Trojan horse detection method based on Trojan horse virus type classification modeling
The invention discloses a Trojan horse detection method based on Trojan horse virus type classification modeling. The method comprises following steps of (1) classifying found Trojan horse according to characteristics; (2) forming a Trojan horse identification class library; (3) collecting characteristics of an operation system, identifying the Trojan horse through the Trojan horse identification class library in the step (2), and positioning in the belonging categories and characteristics of Trojan horse in; (4) positioning suspicious items; (5) according to the collected characteristics of the operation system in the step (3), conducting pattern matching in the class library through an algorithm to identify the Trojan horse in the system; (6) finding the same Trojan horse with pattern matching in the class library, and judging the Trojan horse to be detected. The method can conduct rapidly identification and analysis for existing Trojan horse in the system and particularly for the unknown novel Trojan horse. Compared with a traditional detection manner, detection capability for the Trojan horse, particularly for recognition and detection capability for the unknown novel Trojan horse has great improvement.
Owner:SICHUAN CINGHOO TECH
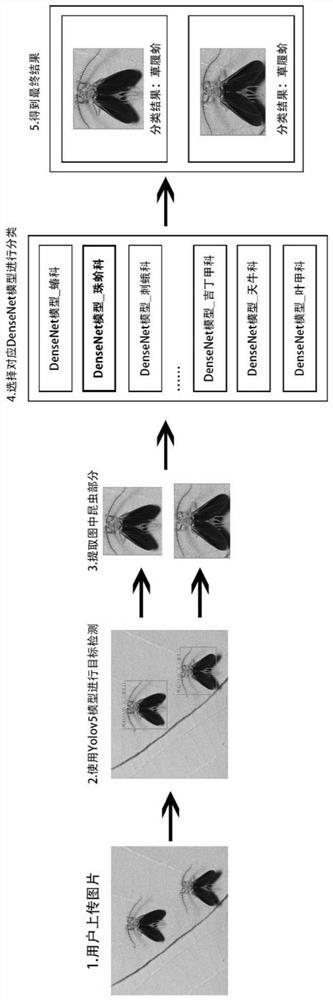
Web online species recognition method based on blade complete and partial two-value characteristics
InactiveCN103824083ADesign specificationEasy maintenanceCharacter and pattern recognitionState of artWeb service
The invention relates to a Web online species recognition method based on blade complete and partial two-value characteristics. The method includes the following steps of firstly, uploading blade images and interaction operation information to a Web server by means of a browser through an uploading interface; secondly, obtaining the blade images through the Web server, and conducting image preprocessing on the blade images; thirdly, extracting new complete and partial two-value textural features of the blade images in a new complete and partial two-value mode; fourthly, enabling the extracted new complete and partial two-value textural characteristics to serve as input of an SVM classifier, recognizing the blade images, and obtaining the serial numbers of species classifications corresponding to the blade images; fifthly, feeding the serial numbers of the species classifications and corresponding species information back to the browser through the Web server. Compared with the prior art, the method has the advantages of being high in accuracy, convenient and rapid to implement and the like.
Owner:TONGJI UNIV
Method and device for modifying disease description
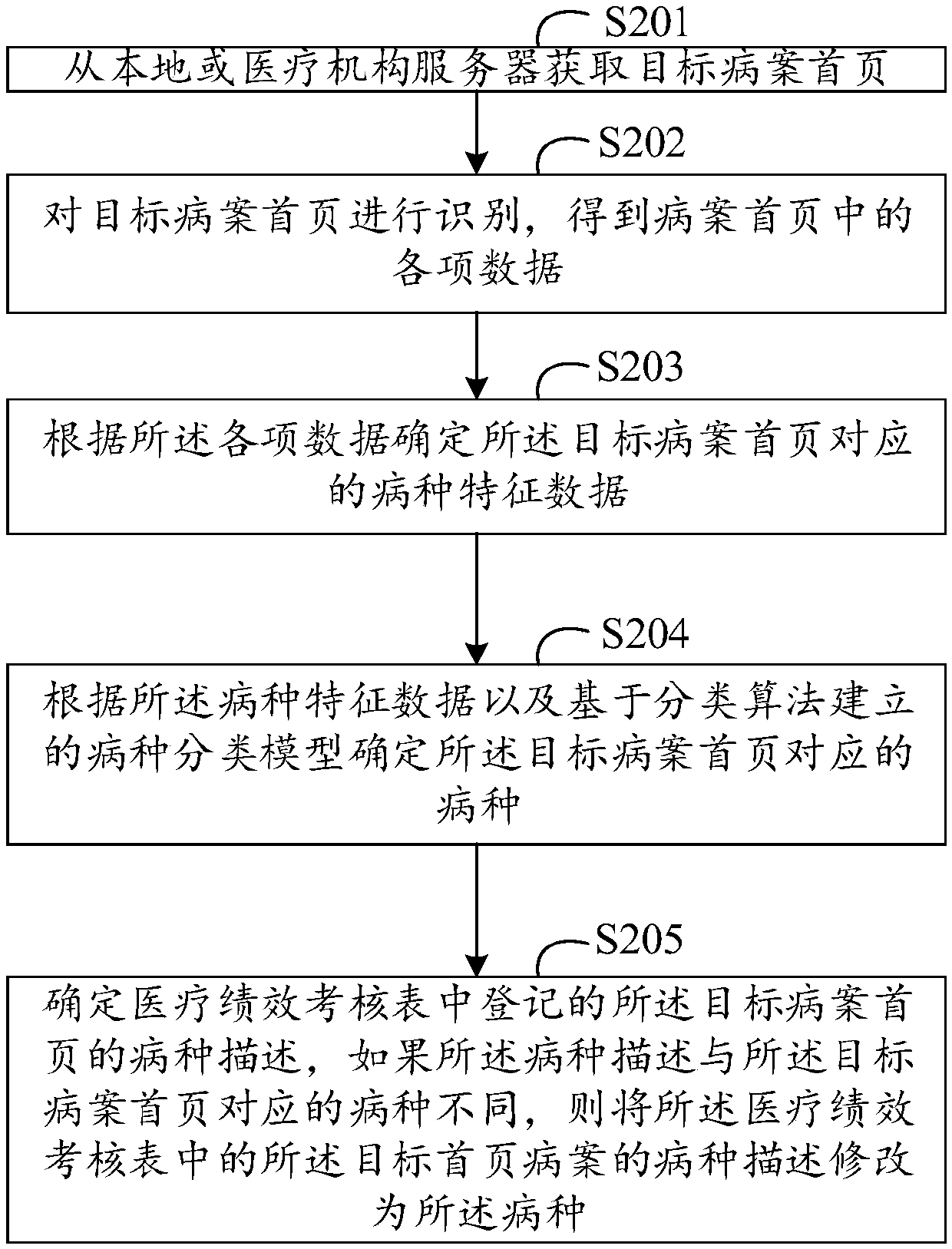
PendingCN109543718AAchieve correctionFacilitate performance analysisMedical data miningCharacter and pattern recognitionMedical recordHome page
The invention provides a method and a device for modifying disease description, wherein, the method comprises: obtaining a target medical record homepage from a local or medical institution server; Identifying the front page of the target medical record to obtain various data in the front page of the medical record; Determining disease type characteristic data corresponding to the front page of the target medical record according to the data; Determining the disease species corresponding to the front page of the target medical record according to the disease species characteristic data and thedisease species classification model established based on the classification algorithm; Determining a disease description of the front page of the target medical record registered in the medical performance appraisal form, and modifying the disease description of the front page of the target medical record in the medical performance appraisal form to the disease type if the disease description isdifferent from the disease type corresponding to the front page of the target medical record. The technical proposal of the invention can correct the nonstandard description of the disease type in the front page of the medical record and facilitate the performance analysis according to the disease type corresponding to the front page of the medical record.
Owner:PING AN MEDICAL & HEALTHCARE MANAGEMENT CO LTD
Pathogenic microorganism metagenome detection method based on third-generation sequencing
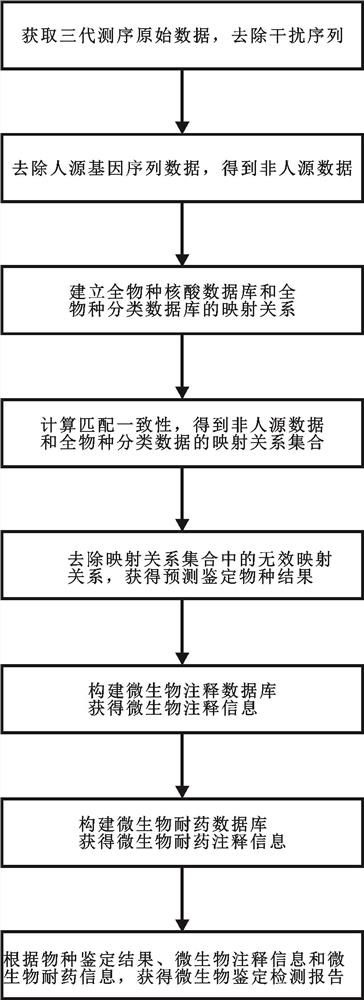
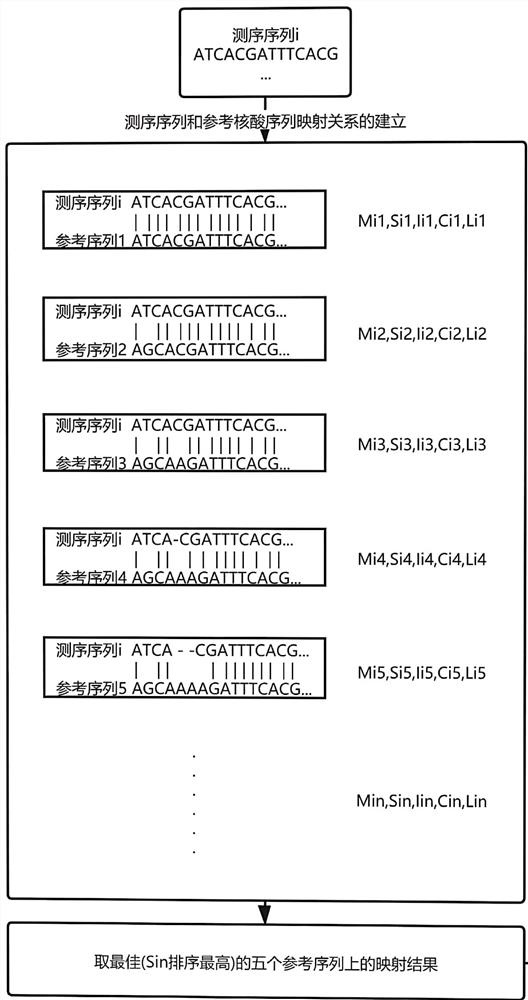
ActiveCN113160882AImprove accuracyOptimizing Mapping ResultsProteomicsGenomicsPathogenic microorganismSpecies classification
The invention discloses a pathogenic microorganism metagenome detection method based on third-generation sequencing, the method comprises the following steps: acquiring original gene detection data of third-generation sequencing of a sample, removing interference sequences, and retaining non-human data; establishing a mapping relationship between the whole species nucleic acid database and the whole species classification database; removing invalid mapping relationships in the mapping relationship set to obtain an effective mapping relationship set of the non-human source data and the whole species classification database, and performing calculation according to the effective mapping relationship set to obtain a species identification result; constructing a microorganism annotation database to obtain microorganism annotation information; constructing a microbial drug resistance database to obtain microbial drug resistance information; and obtaining a microorganism detection report according to the species identification result, the microorganism annotation information and the microorganism drug resistance information. According to the method, the non-human source data is established, the mapping result is optimized, the detection precision is improved, the accuracy of species classification in similar regions is improved, and then species are obtained through prediction and comparison of sequence results.
Owner:成都博欣医学检验实验室有限公司
Method and device for detecting pathogenic microorganisms based on metagenomics
ActiveCN113744807AExpand the scope ofImprove accuracySequence analysisInstrumentsReference genesGenomics
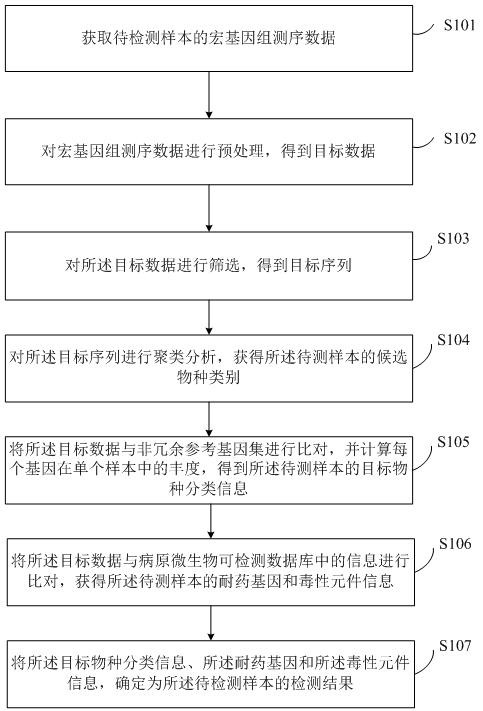
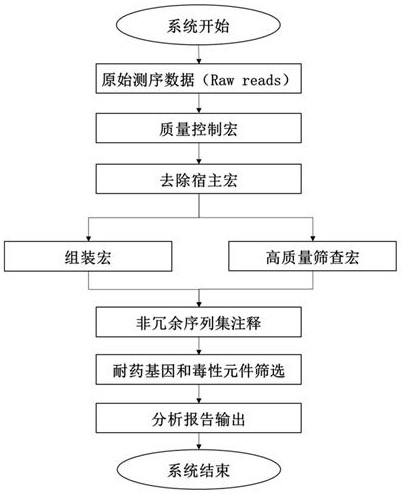
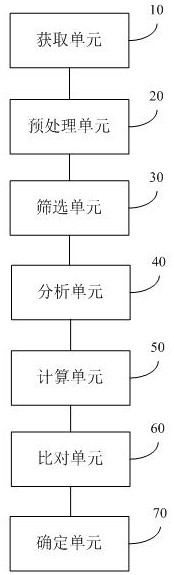
The invention discloses a pathogenic microorganism detection method and device based on metagenomics. The method comprises the following steps: acquiring metagenome sequencing data of a to-be-detected sample; preprocessing the metagenome sequencing data to obtain target data; screening the target data to obtain a target sequence; performing clustering analysis on the target sequence to obtain candidate species categories of the to-be-detected sample; comparing the target data with the non-redundant reference gene set, and calculating abundance of each gene in a single sample to obtain target species classification information of the to-be-detected sample; comparing the target data with information in a pathogenic microorganism detectable database to obtain drug-resistant gene and toxic element information of the to-be-detected sample; and determining the target species classification information, the drug-resistant gene and the toxic element information as a detection result of the to-be-detected sample. According to the invention, the pathogenic microorganism detection applicability range and the pathogenic detection accuracy are improved.
Owner:微岩医学科技(北京)有限公司 +1
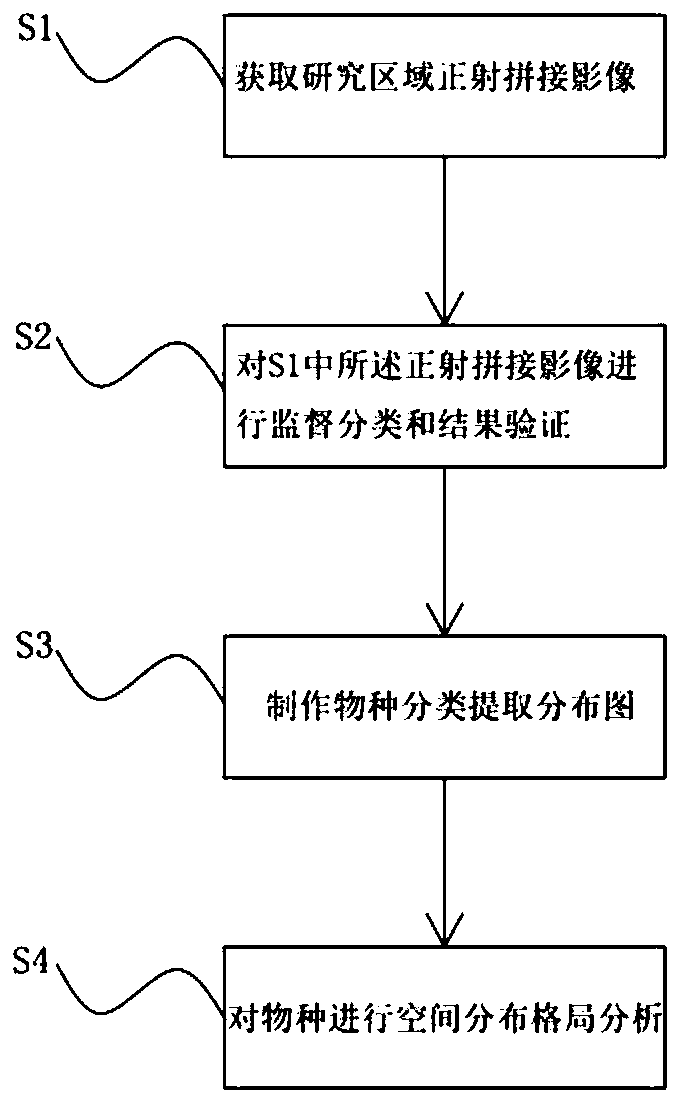
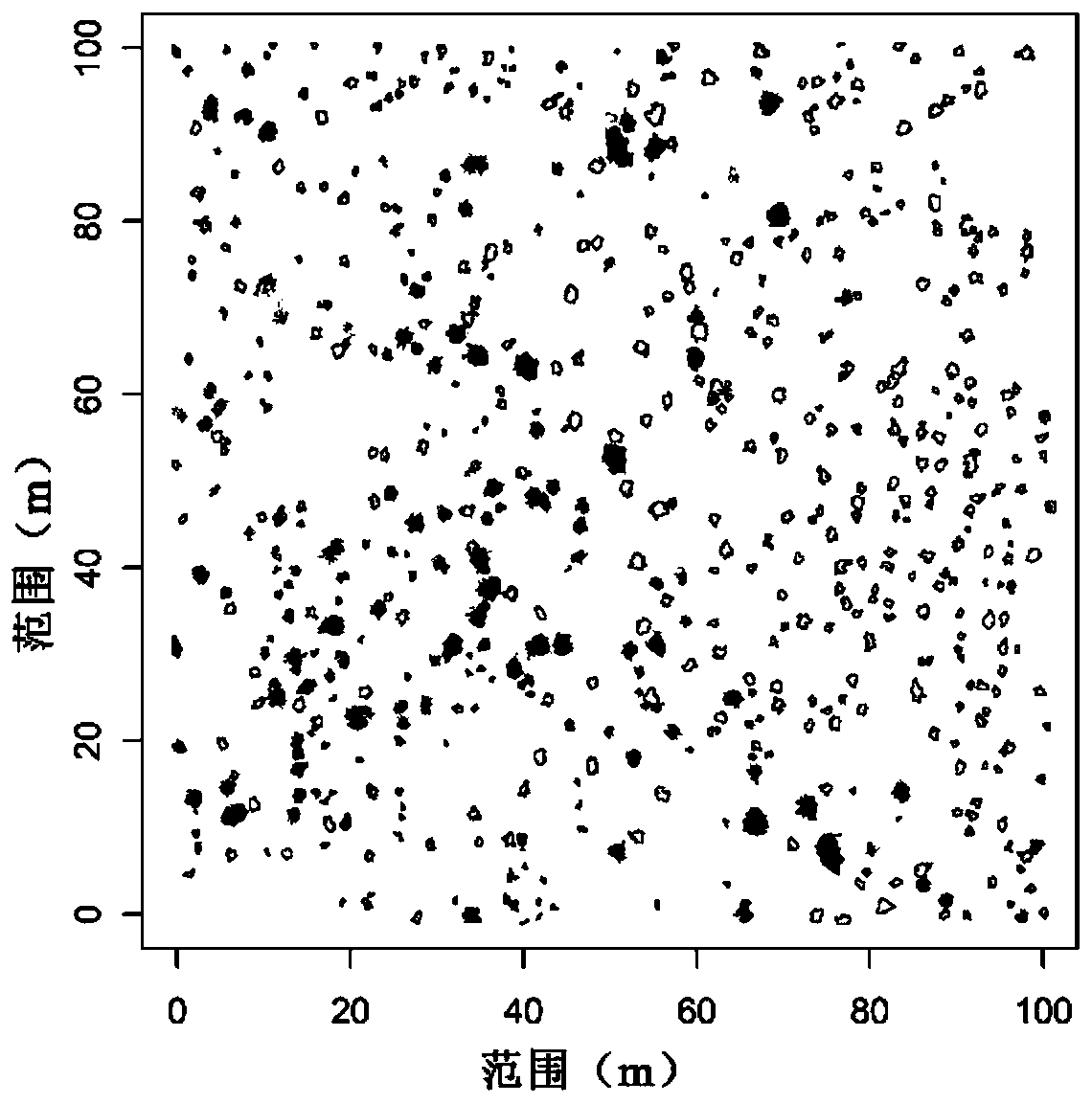
Vegetation spatial distribution pattern investigation method and vegetation classification method based on unmanned aerial vehicle technology
PendingCN111444824ASave manpower and material resourcesComprehensive and Accurate AnalysisGeometric image transformationCharacter and pattern recognitionStatistical analysisSpecies classification
The invention discloses a vegetation spatial distribution pattern investigation method based on an unmanned aerial vehicle technology. The method comprises the following steps: S1, obtaining an orthographic spliced image of a research area; S2, performing supervised classification and result verification on the orthographic spliced image in S1; S3, making a species classification extraction distribution diagram; and S4, performing spatial distribution pattern analysis on the species. The method has the beneficial effects that vegetation species classification is carried out on the orthographicspliced images by utilizing a supervised classification method, so that specific ecological statistical analysis is carried out on individuals of each species; manpower and material resources are saved, meanwhile, a pattern analysis quadrat can be expanded by tens of times, and a spatial pattern analysis conclusion can be obtained more comprehensively and accurately; multi-angle spatial distribution pattern analysis can be more conveniently carried out on the species classification extraction distribution diagram, such as a sample method, a point pattern analysis method and the like, so thatpattern analysis results are more comprehensive and specific.
Owner:PEKING UNIV SHENZHEN GRADUATE SCHOOL
Amplification primer and method of nassariidae mitochondria COI gene
InactiveCN104388571AEfficient amplificationThe amplification efficiency of the amplification primers is effectiveMicrobiological testing/measurementDNA/RNA fragmentationSpecies classificationToxin
The invention belongs to the field of molecular biology, and provides an amplification primer of a nassariidae mitochondria COI gene, and a method for amplifying the nassariidae mitochondria COI gene by using the primer. According to the amplification primer of the nassariidae mitochondria COI gene, the sequences are ZLCO1490F:5'-TTTCTACAAATCATAAAGACATTGG-3' and ZHCO2198R:5'-TATACCTCCGGGTGACCAAAAAATCA-3'. The amplification primer of the nassariidae mitochondria COI gene provided by the invention is capable of effectively amplifying COI sequences of different species of the nassariidae, and has great significance in research of species classification, system development, systematic geography, toxin tolerance and protection of nassariidae genetic resources by using the COI sequences.
Owner:NANJING AGRICULTURAL UNIVERSITY
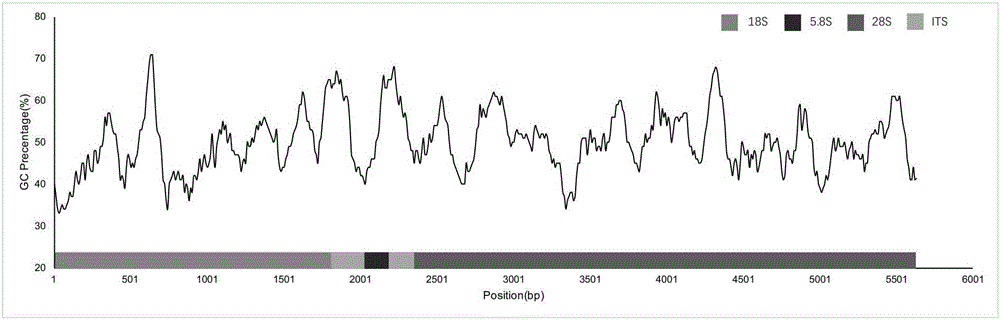
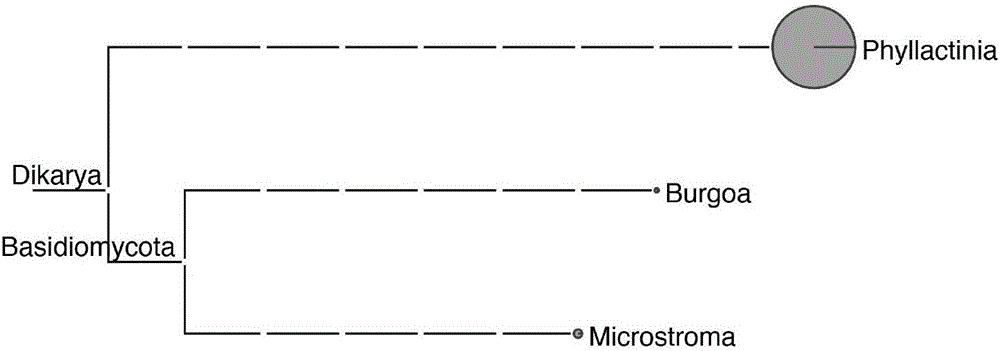
Mulberry powdery mildew pathogenic bacteria Phyllactinia mori ribosome RNA sequence and application thereof
ActiveCN106801053AMicrobiological testing/measurementDNA/RNA fragmentationSpecies classificationFull length cdna
The invention discloses a full-length cDNA sequence of mulberry powdery mildew pathogenic bacteria Phyllactinia mori ribosome RNA and application thereof. The full-length cDNA sequence of the mulberry powdery mildew pathogenic bacteria Phyllactinia mori ribosome RNA is shown in SEQ.ID.NO 1, and two pairs of primer sequences contained in the full-length cDNA sequence are shown as SEQ.ID.NO 2 to 5. According to the full-length cDNA sequence of the mulberry powdery mildew pathogenic bacteria Phyllactinia mori ribosome RNA, the cDNA sequence of the Phyllactinia mori ribosome RNA is applied to detecting Phyllactinia mori bacteria, and qualitative and quantitative results can be obtained at the same time. The results show that fungus with the highest relative abundance in study on fungus on mulberry powdery mildew leaves is Phyllactinia mori pathogenic bacteria. In addition, the cDNA sequence of the Phyllactinia mori ribosome RNA can be applied to study on fungus species classification.
Owner:SOUTH CHINA AGRI UNIV
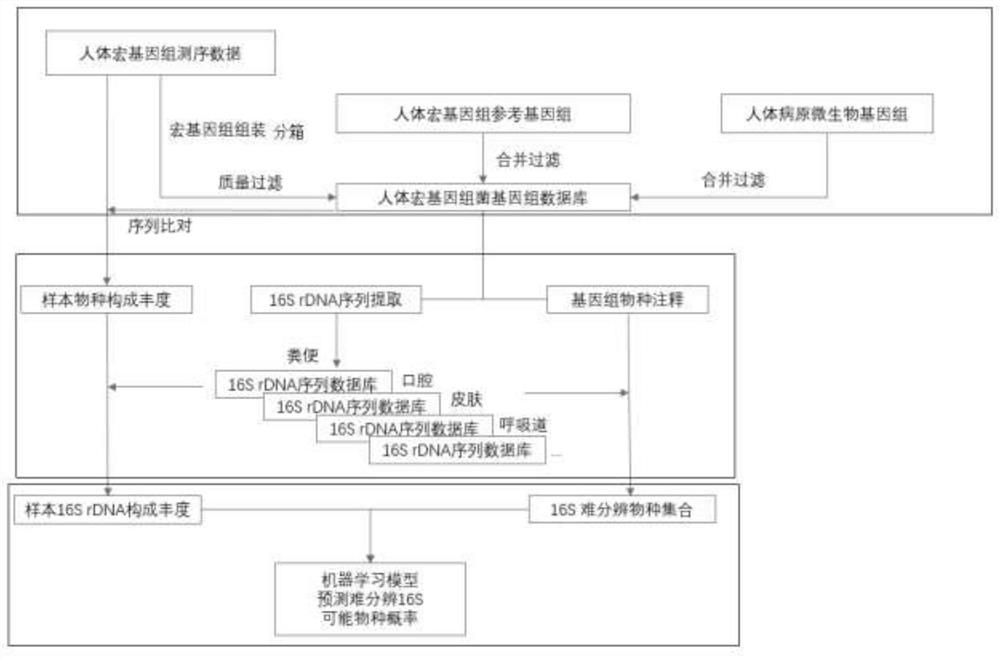
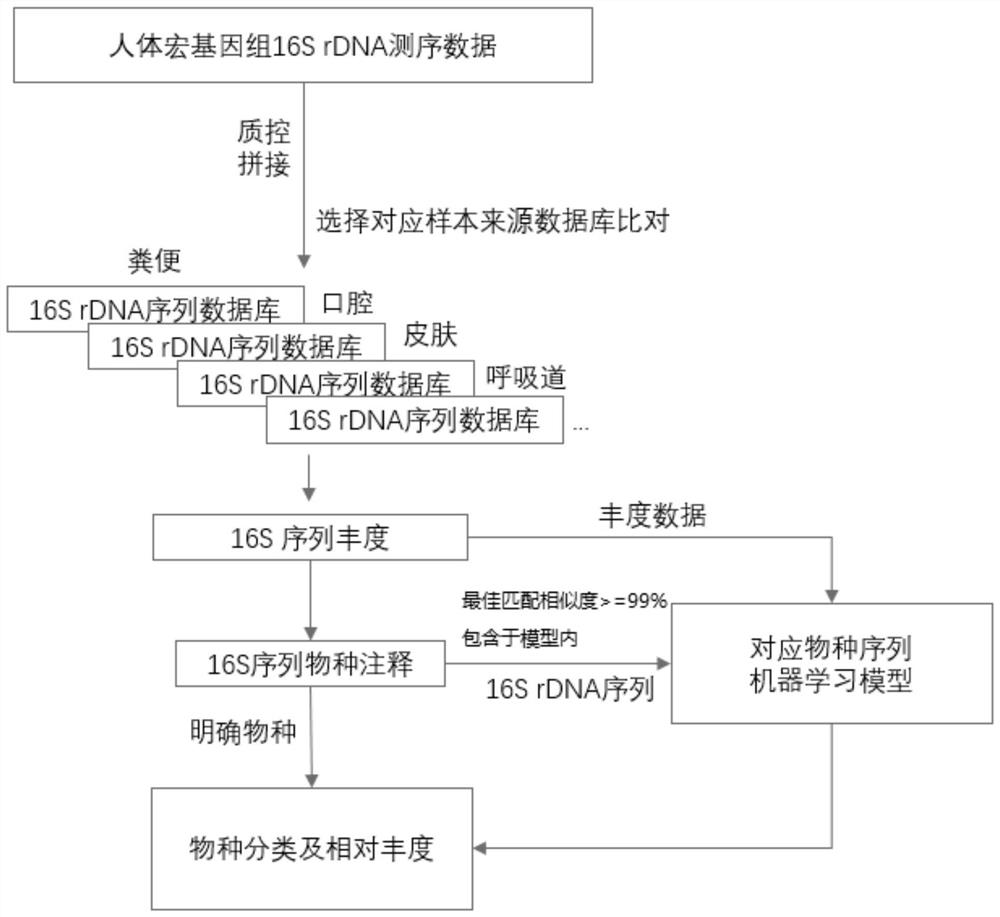
Optimization method for accurate species identification of human flora through high-throughput 16S rDNA sequencing
PendingCN111816258AReduce similar sequence interferenceImprove comparison efficiencyBiostatisticsSequence analysisMicroorganismGenomic databases
The invention discloses an optimization method for accurate species identification of human flora through high-throughput 16S rDNA sequencing. The optimization method comprises the following steps: 1)establishing a human metagenome bacterium genome database based on human microorganism samples; 2) extracting full-length 16S rDNA sequences of the human metagenomic bacterium genome database, and establishing 16S rDNA species annotation databases of different human body parts; and 3) establishing a sample-strain 16S sequence abundance correlation network, and training a machine learning model byusing the data. The invention also provides a method for performing species identification and abundance calculation on the high-throughput sequencing 16S rDNA sequences by using the constructed database and model. According to the invention, the species classification resolution and accuracy of 16S rDNA can be greatly improved, so an analyst can find more accurate and clear strains according toresults.
Owner:HANGZHOU GUHE INFORMATION TECH CO LTD
Amphibian mitochondria general macro bar code amplification primers and application method thereof
PendingCN111549146AImprove general performanceGuaranteed resolutionMicrobiological testing/measurementDNA/RNA fragmentationForward primerPhylogenetic relationship
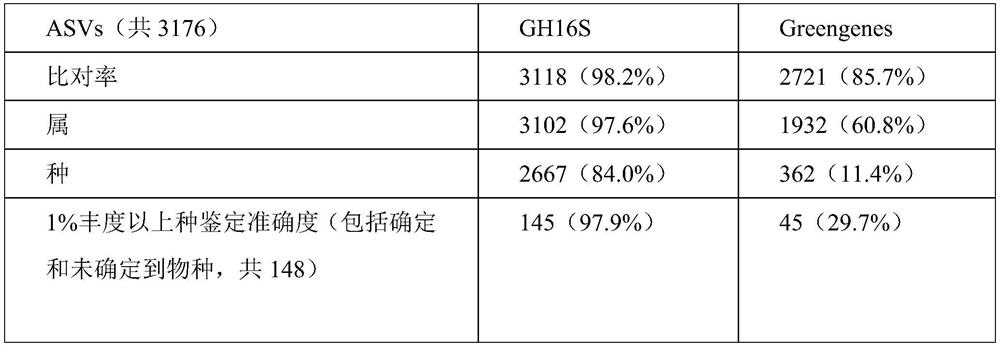
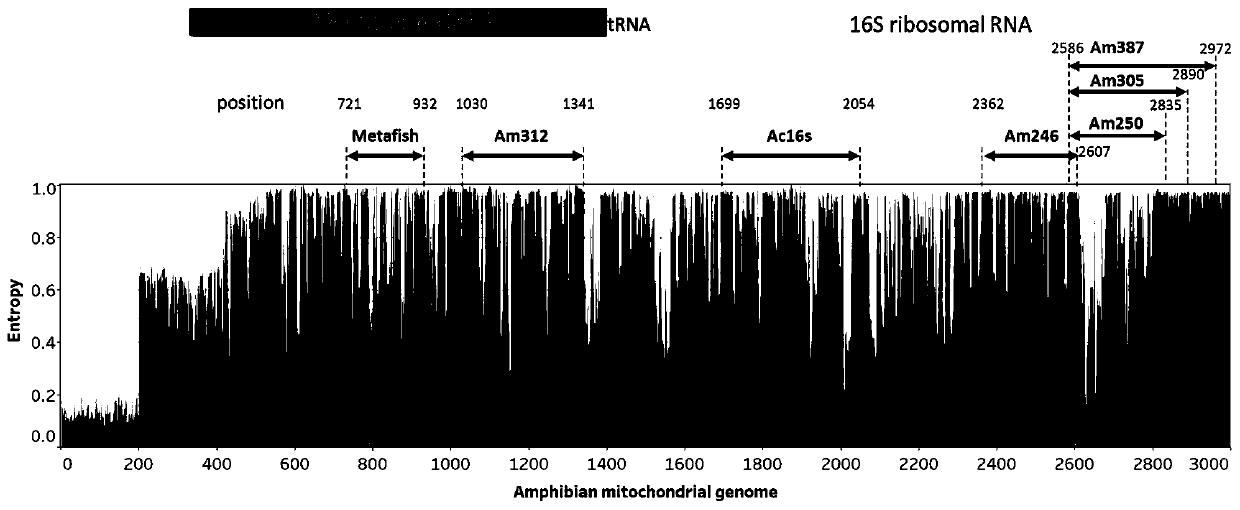
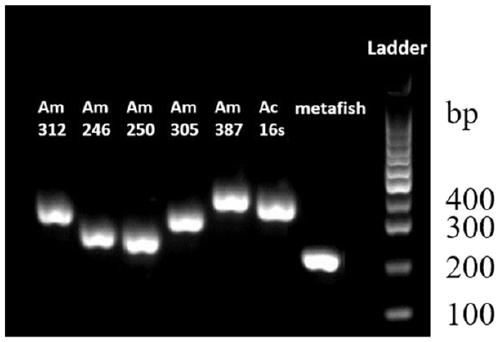
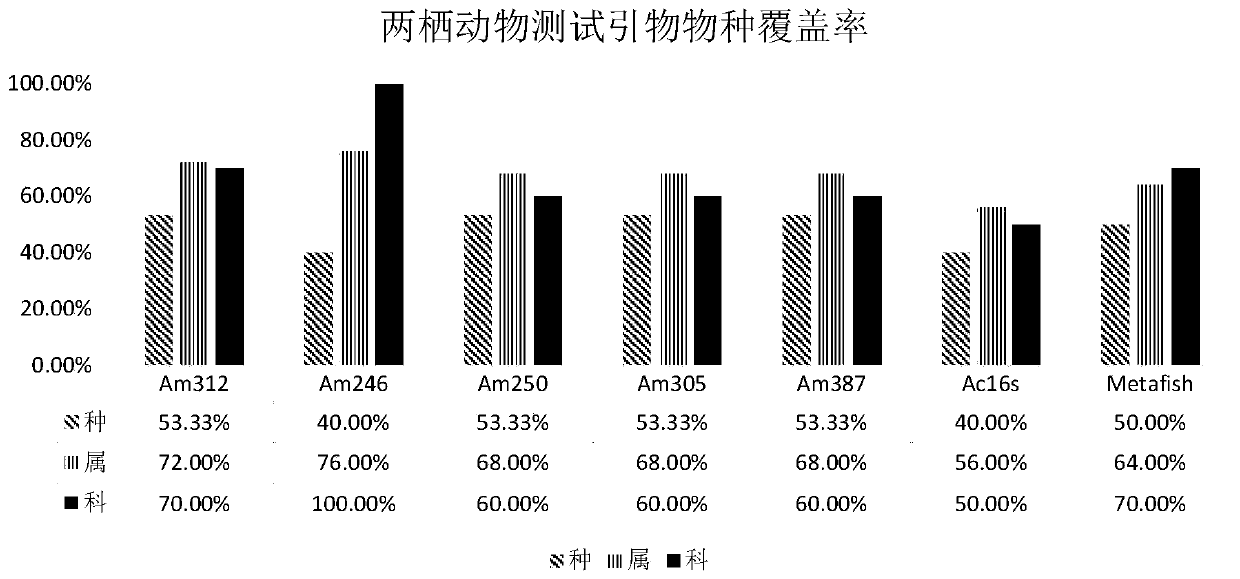
The invention belongs to the technical field of gene detection, and discloses amphibian mitochondria general macro bar code amplification primers and an application method thereof. The amplification primers comprise a primer pair formed by combining at least one forward primer and one reverse primer. Products obtained by PCR amplification through combination of Am312-F1 and Am312-R1, combination of Am246-F2 and Am246-R2, combination of Am250-F3 and Am250-R3.1, combination of Am250-F3 and Am305-R3.2 and combination of Am250-F3 and Am387-R3.3 meet the requirements of next-generation sequencing and can be used for species identification and diversity analysis of amphibians; through flexible combination of the above primer pairs, the research purposes of amphibian species classification, phylogenetic relationship, systematic geography and the like can be achieved.
Owner:南京易基诺环保科技有限公司
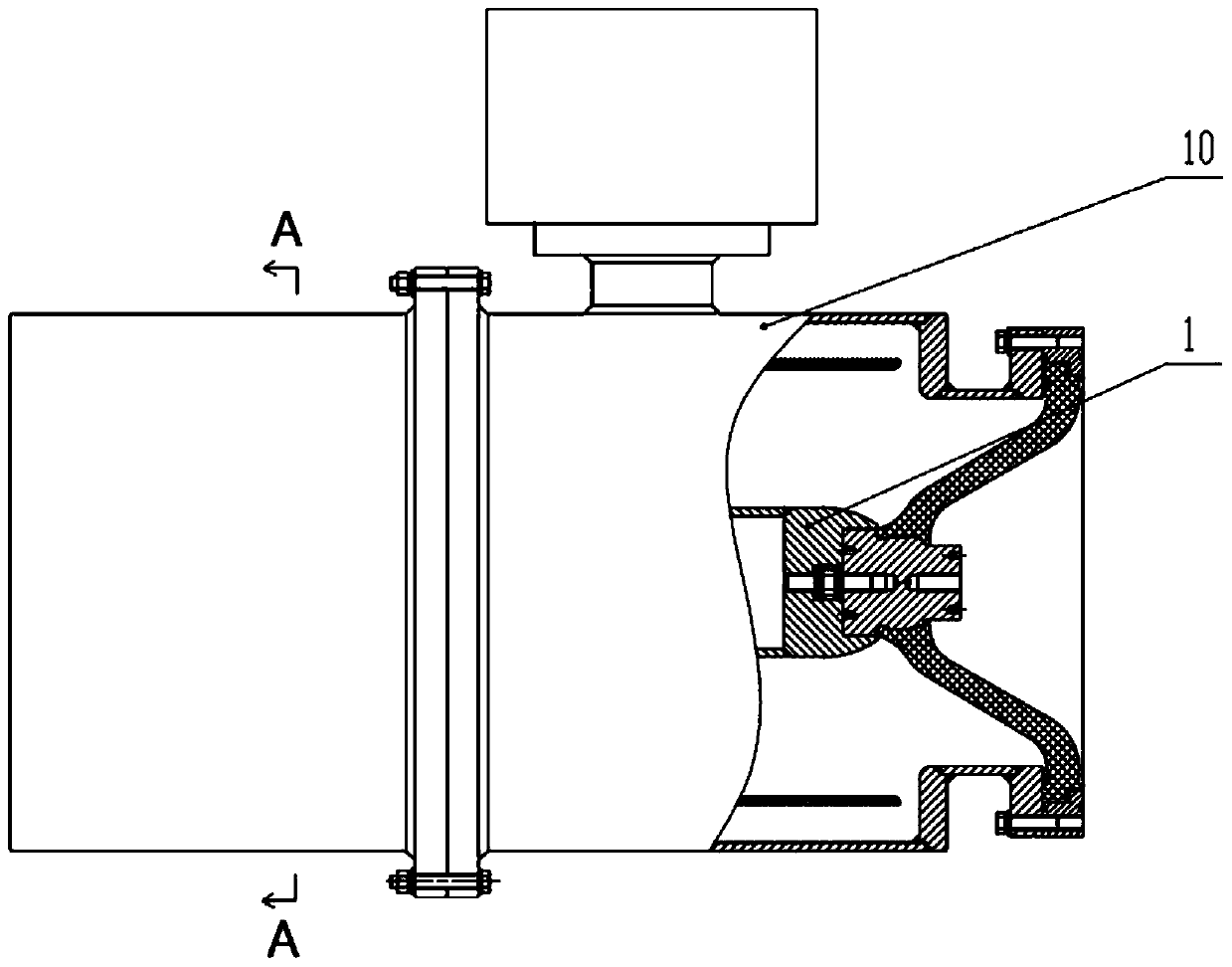
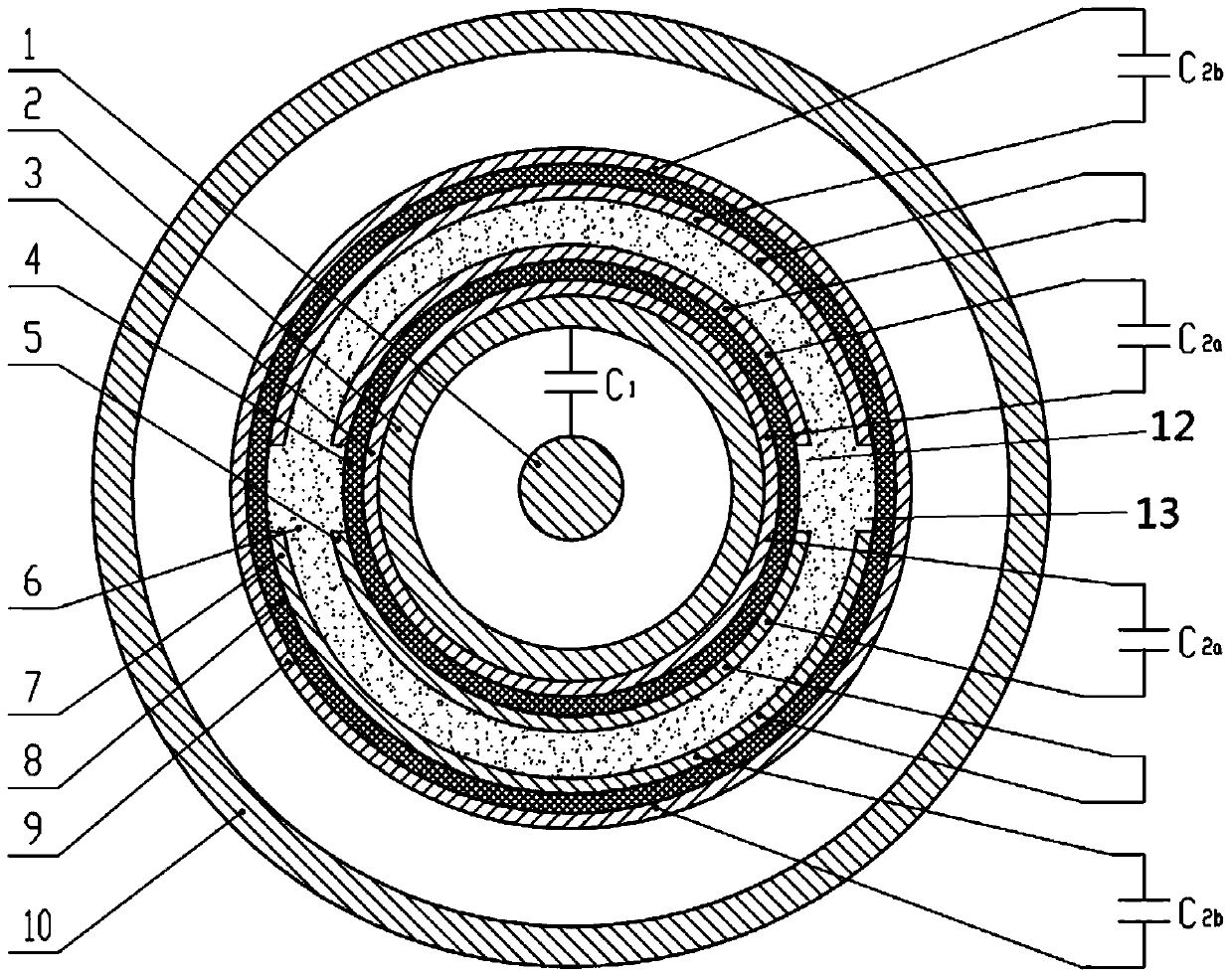
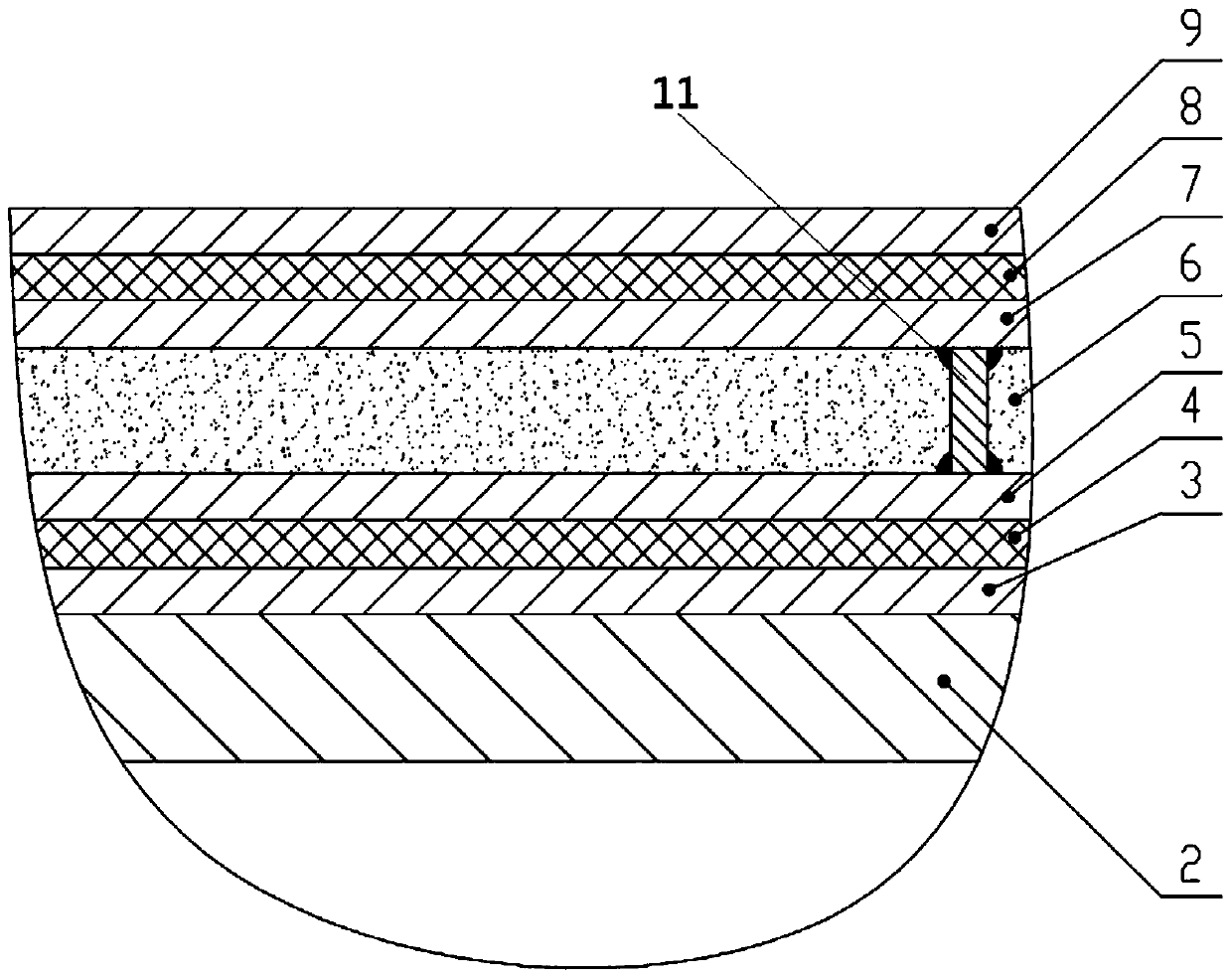
A graded voltage transformer
ActiveCN105467187BEasy to processSimple processTransformersVoltage/current isolationLow voltageTransformer
The invention relates to a graded type voltage transformer comprising an inner insulation dielectric layer and an external insulation dielectric layer which are arranged in a coaxial mode; the interior surface of the inner insulation dielectric layer is bonded with a first metal layer; the external surface of the insulation dielectric layer is bonded with a second metal layer; the interior surface of the external insulation dielectric layer is bonded with a third metal layer positioned on the outer side of the second metal layer; the external surface of the external insulation dielectric layer is bonded with a fourth metal layer; the second metal layer comprises at least one inner side low-voltage electrode; the third metal layer comprises an outer side low-voltage electrode which is the same with the inner side low-voltage electrode in terms of quantity; and the inner side low-voltage electrode is in short circuit with the outer side low-voltage electrode. According to the invention, problems that a continuous arranged structure of a metal layer, an insulation dielectric layer, a metal layer, an insulation dielectric layer and a metal layer has relatively high requirements for processing technology in the prior art are solved.
Owner:STATE GRID CORP OF CHINA +2
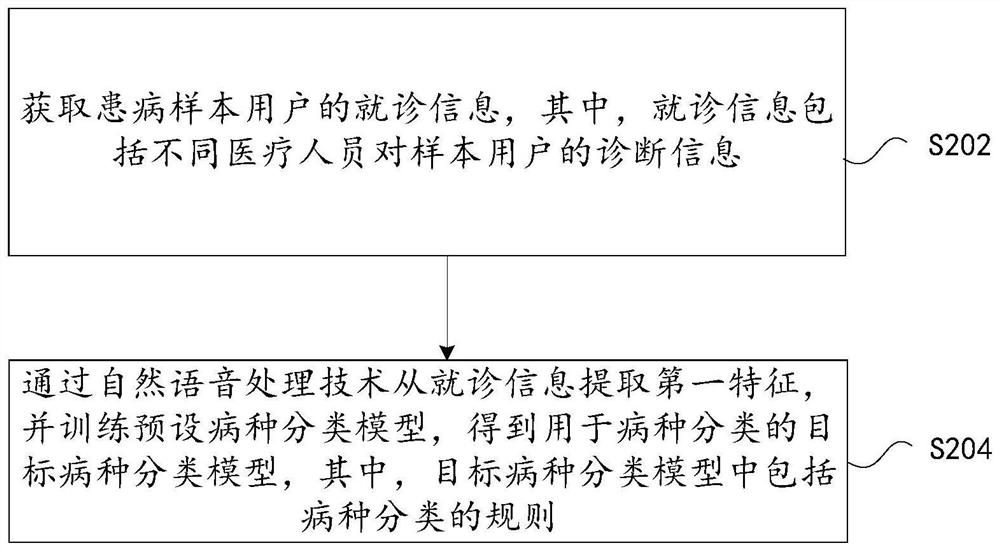
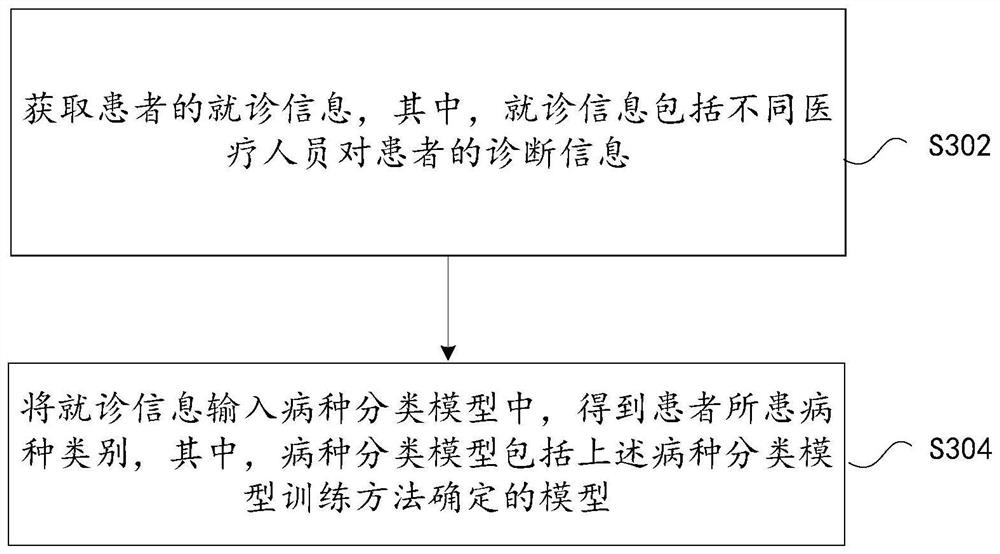
Disease classification model training method and device, storage medium and electronic device
PendingCN114117053AQuick classificationAccurate classificationSpecial data processing applicationsText database clustering/classificationSpecies classificationPhysical therapy
The invention discloses a disease classification model training method and device, a storage medium and an electronic device. The disease category classification model training method comprises the steps that treatment information of a disease sample user is acquired, and the treatment information comprises diagnosis information of different medical personnel on the sample user; a first feature is extracted from the doctor seeing information through a natural speech processing technology, a preset disease classification model is trained, a target disease classification model used for disease classification is obtained, the target disease classification model comprises a disease classification rule, and a first feature is extracted from the doctor seeing information; that is to say, the disease species of the patient can be quickly and accurately classified through the trained disease species classification model, and the technical problem of low classification efficiency of the disease species of the patient in the prior art is solved.
Owner:SHENZHEN UNISOUND INFORMATION TECH CO LTD
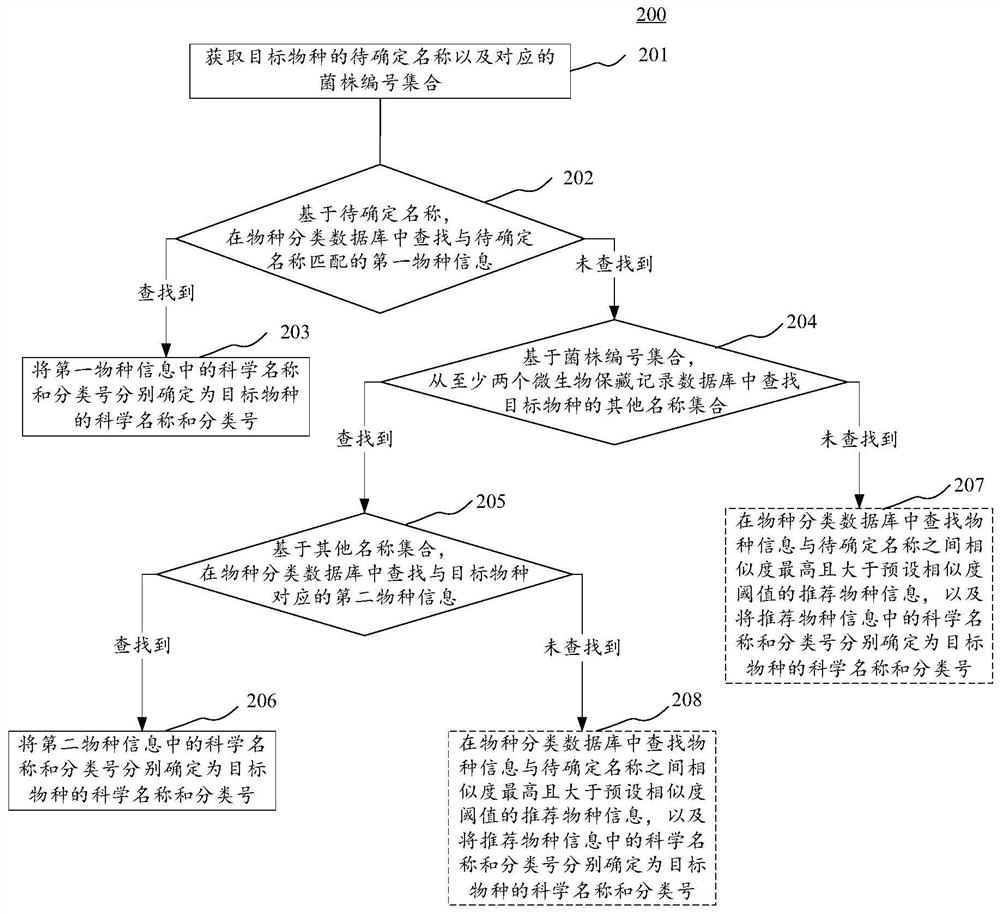
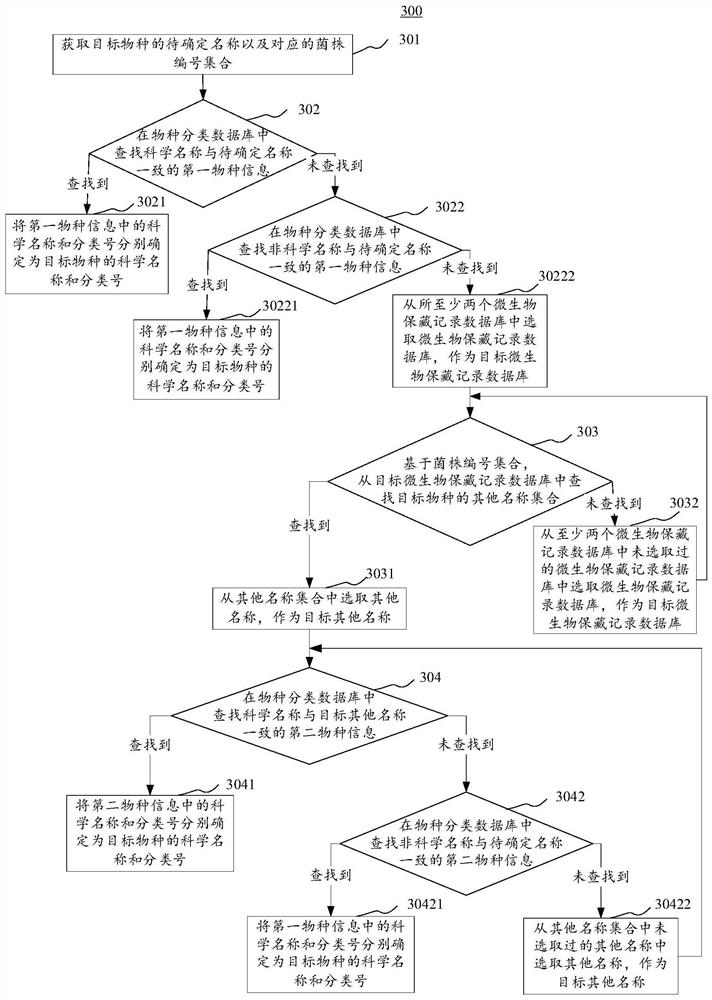
Species science name determination method and device, electronic equipment and storage medium
PendingCN112037865AImprove recallBiostatisticsNatural language data processingMicroorganism preservationMicroorganism
The invention provides a species science name determination method and a device, electronic equipment and a storage medium. One specific embodiment of the method comprises the steps of obtaining a to-be-determined name of a target species and a corresponding strain number set, searching for first species information matched with the to-be-determined name in a species classification database basedon the to-be-determined name, responding to the situation that the first species information matched with the to-be-determined name is not found, and based on the strain number set, searching other name sets of the target species from at least two microorganism preservation record databases, responding to the searched other name sets of the target species, searching second species information corresponding to the target species from a species classification database based on the other name sets, and determining the scientific name and the classification number in the second species informationas the scientific name and the classification number of the target species respectively. According to the embodiment, the recall ratio of species science names is improved.
Owner:INST OF MICROBIOLOGY - CHINESE ACAD OF SCI
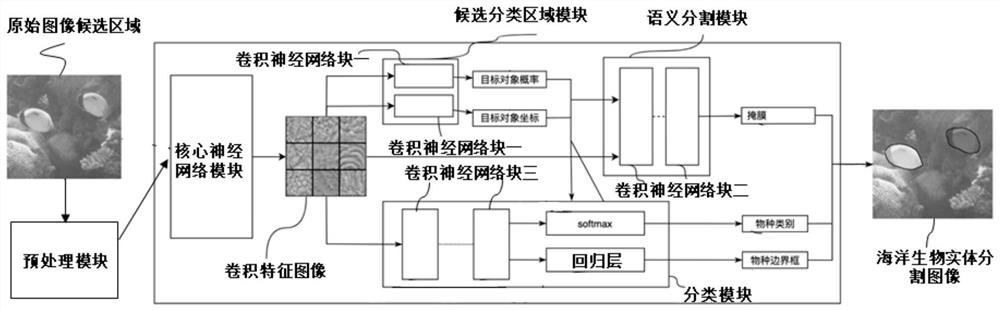
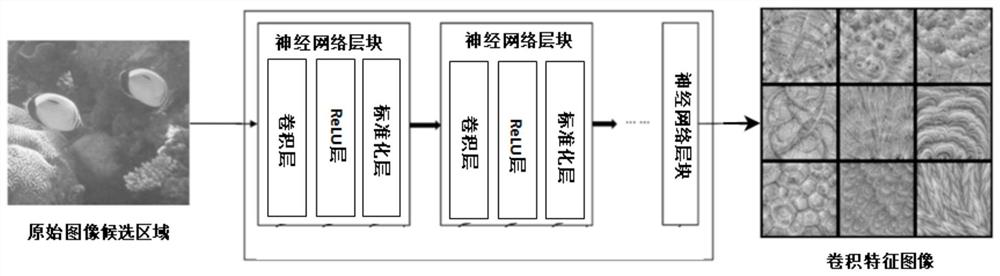
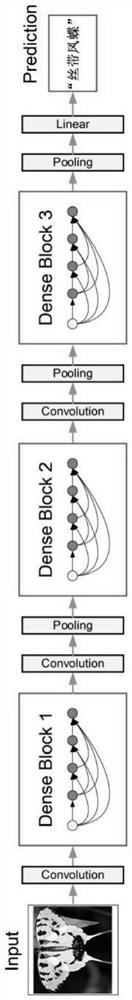
Marine organism identification system and identification method based on deep neural network
PendingCN112149612AResolve identifiabilitySolve the segmentation problemCharacter and pattern recognitionNeural architecturesSpecies classificationImage synthesis
The invention provides a marine organism recognition system based on a deep neural network, and the system comprises a preprocessing module which collects an original image candidate region containinga target object in an original image of a marine organism, and inputs the original image candidate region into the deep neural network DNN; a deep neural network DNN; and an image synthesis module used for carrying out synthesis processing on the species classification, the species boundary frame and mask information and the preprocessed marine organism image to obtain a marine organism entity segmentation image. According to the method, original image information is collected and marked, then calculation processing is carried out in a deep neural network DNN, then marine organisms are recognized, segmented and tracked, and finally the marine organisms are combined with an original image to form visual picture information. In addition, the invention further provides an identification method based on the identification system, and the method effectively solves the technical problem that in the prior art, sea low species cannot be accurately identified, segmented and tracked.
Owner:海略(连云港)科技有限公司
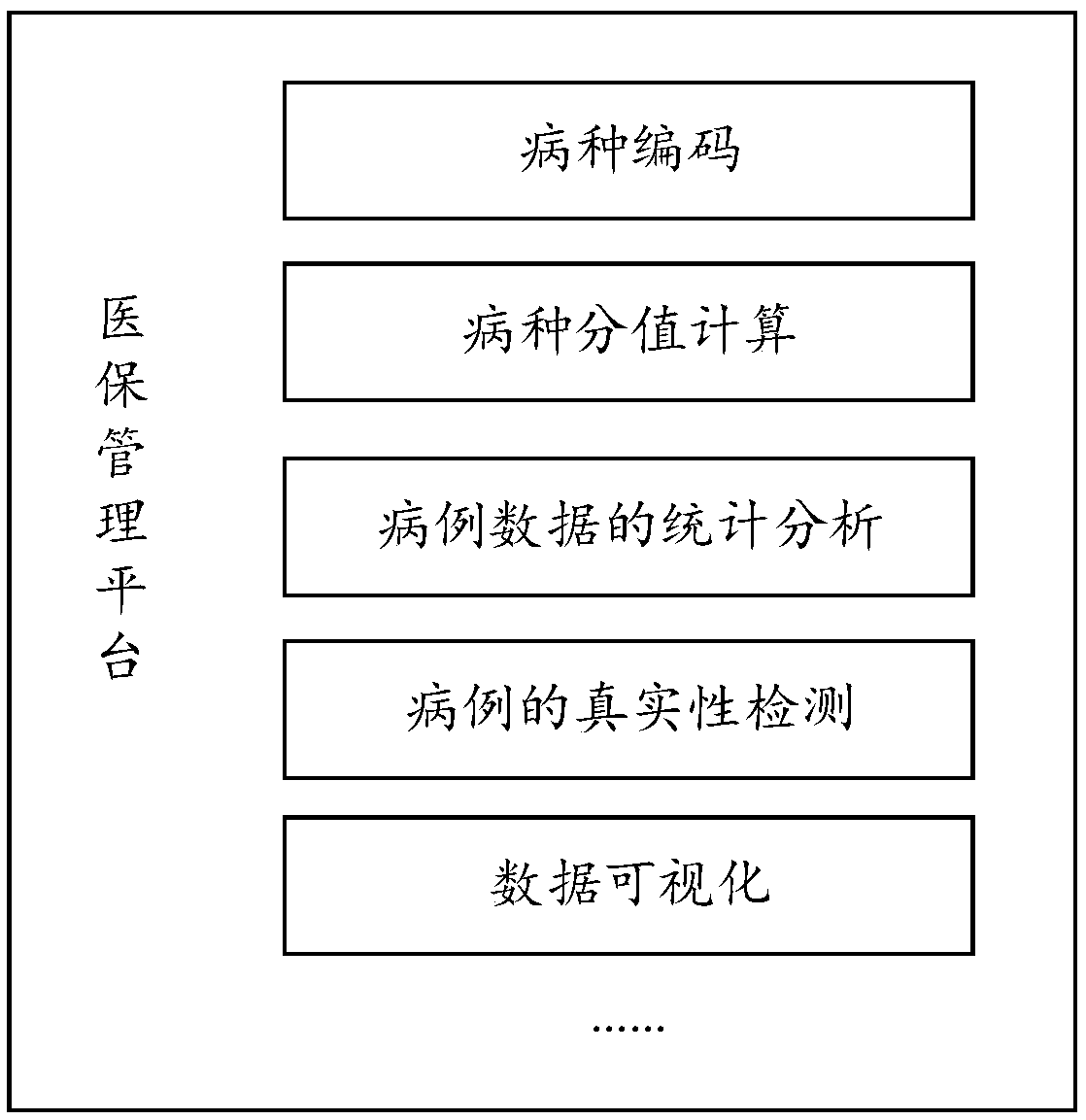
A disease score adjustment based on big data method and a computing device
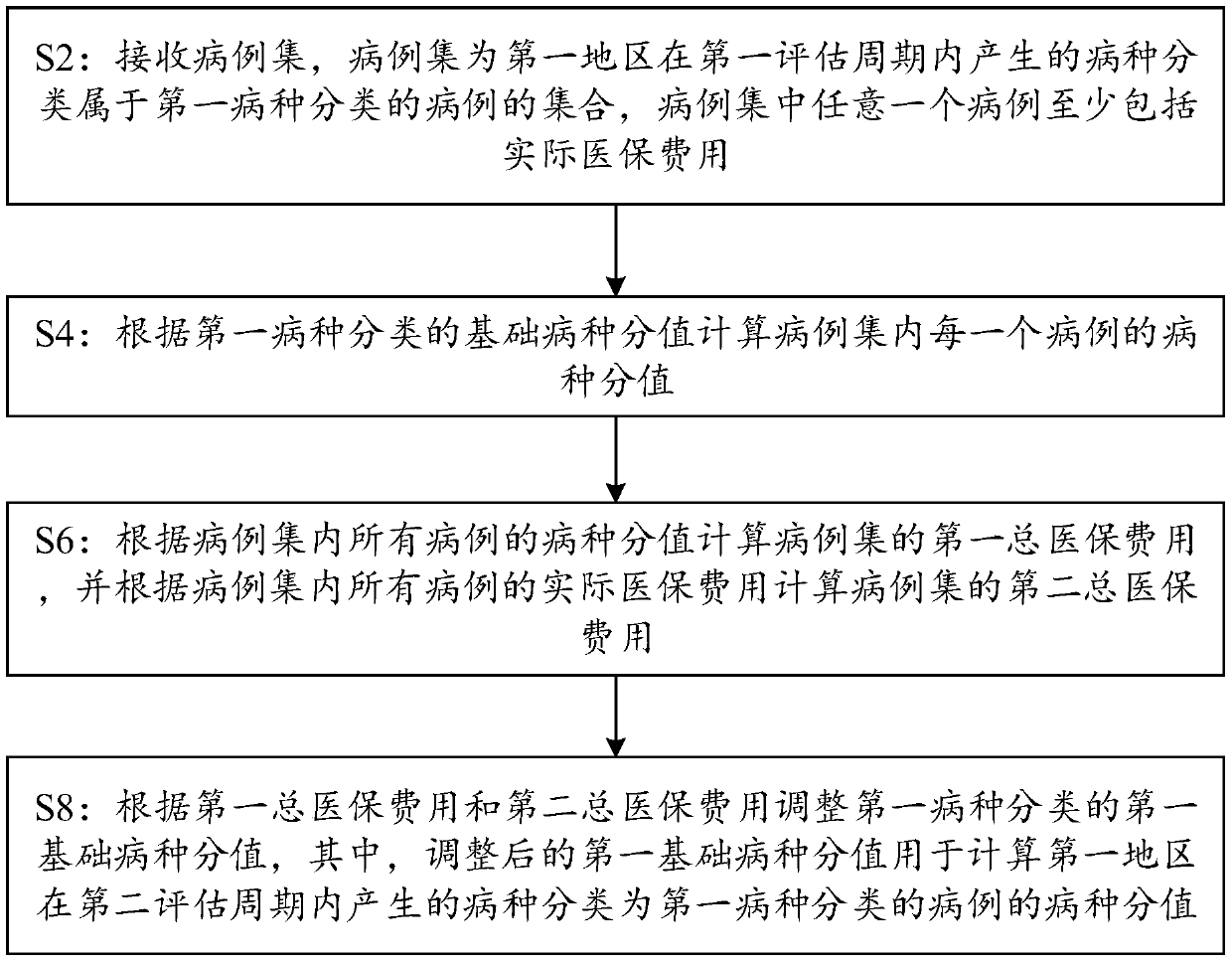
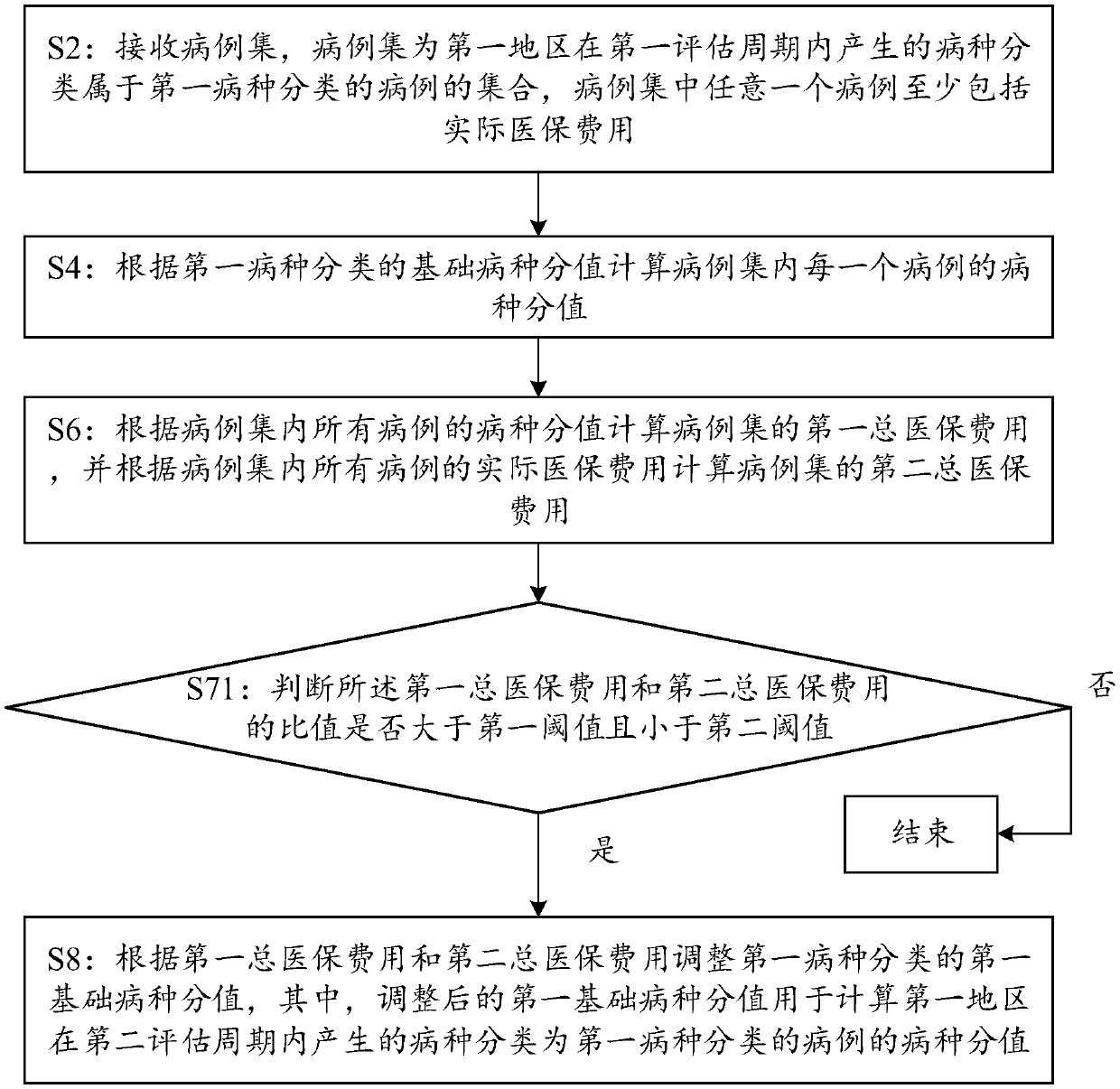
PendingCN109544374AImprove accuracyAchieve regulationFinanceStructured data retrievalSpecies classificationDisease category
The embodiment of the invention discloses a disease score adjustment based on big data method and a computing device. The method comprises the following steps: a computing device receives a case set,the case set is a set of cases generated in a first evaluation cycle in a first region and belonging to a first disease classification, and the case set includes actual medical insurance cost; Calculating a disease species score of each case in the case set according to the basic disease species score of the first disease species classification; Calculating a first total medical insurance cost ofthe case set based on the disease category scores of all cases in the case set, and calculating a second total medical insurance cost of the case set based on the actual medical insurance cost of allcases in the case set; Furthermore, the first basic disease score of the first disease classification is adjusted according to the first total medical insurance cost and the second total medical insurance cost, so as to calculate the disease score of the cases belonging to the first disease classification in the second evaluation cycle by the adjusted first basic disease score, thereby improving the accuracy of the disease score.
Owner:PING AN MEDICAL & HEALTHCARE MANAGEMENT CO LTD
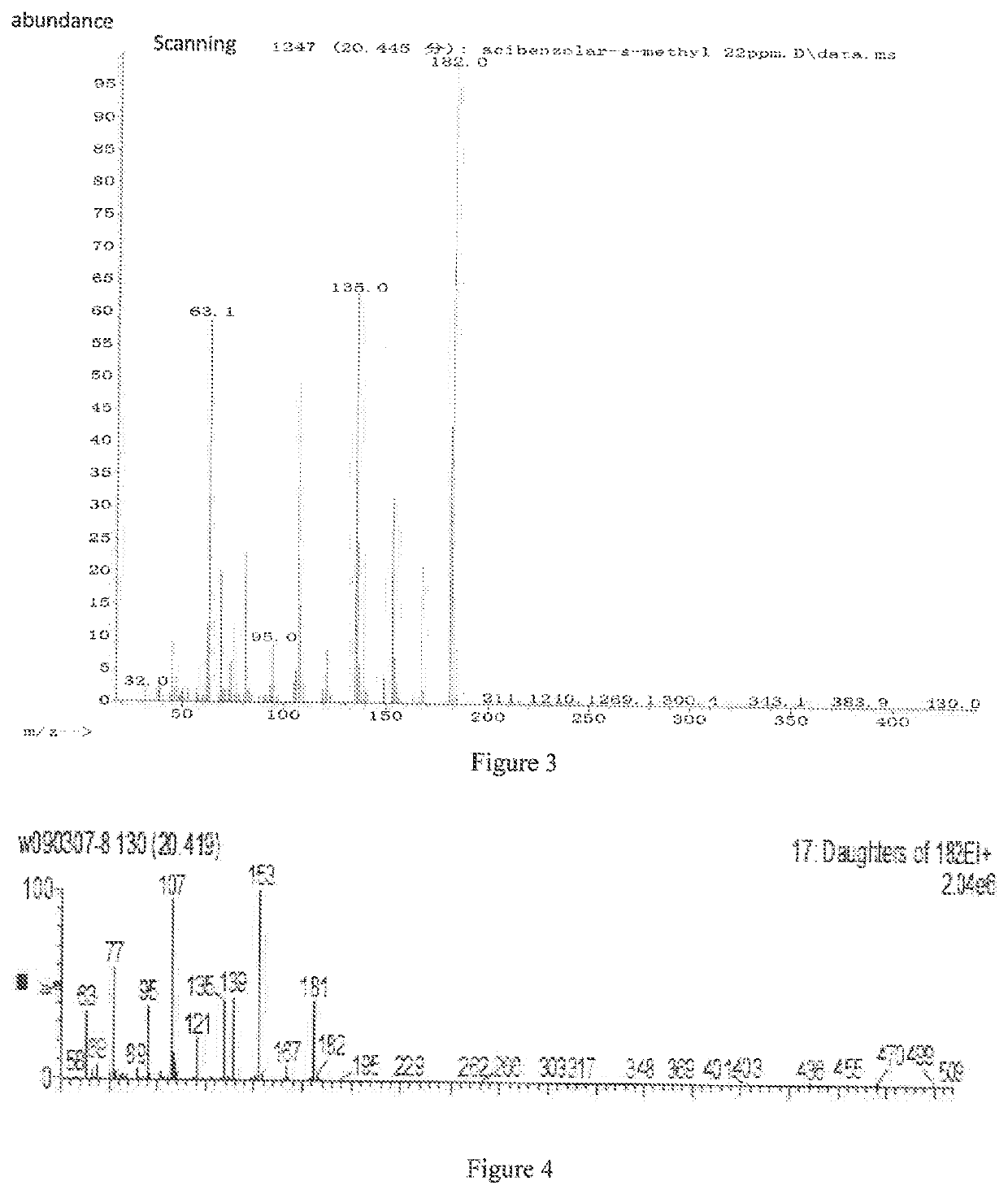
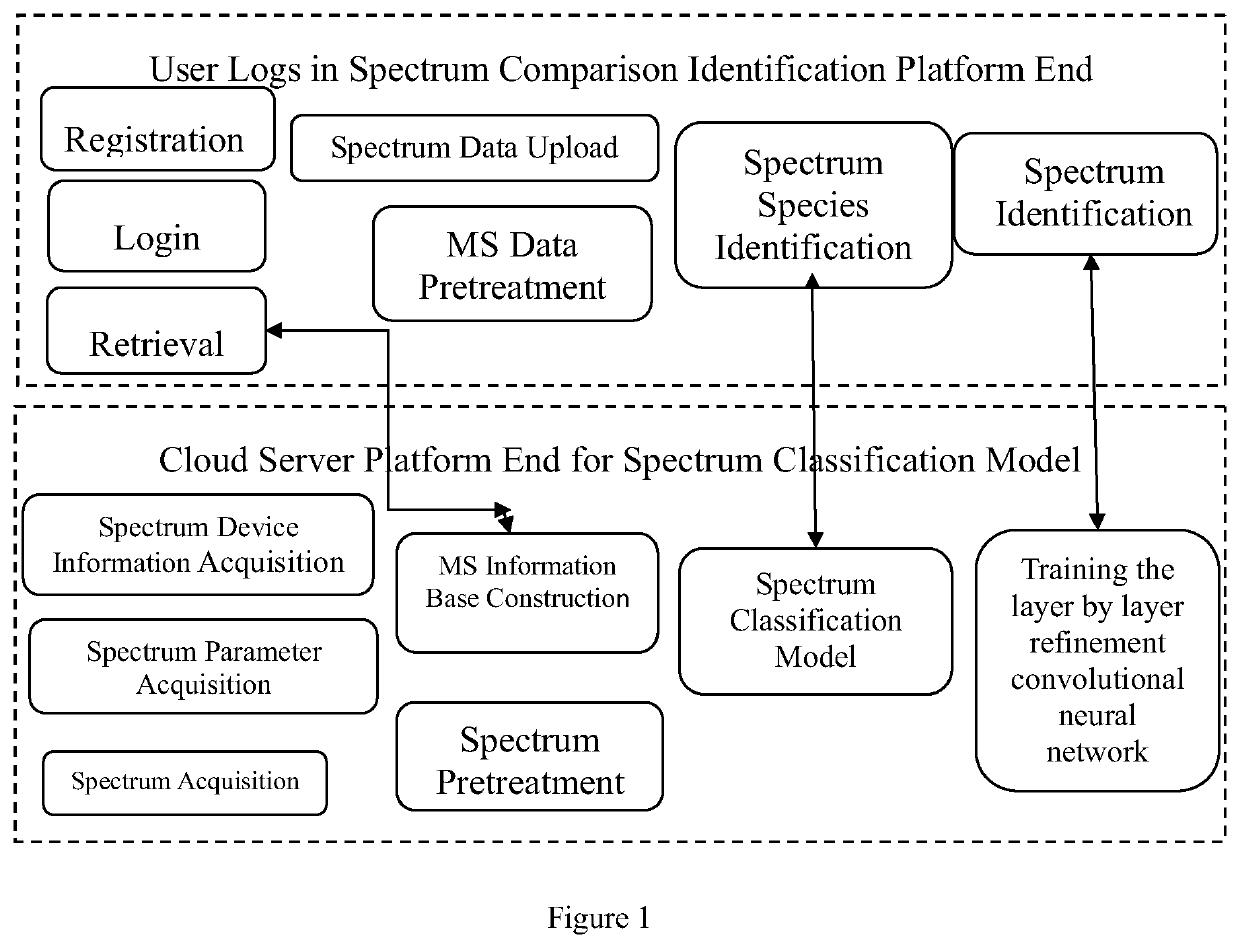
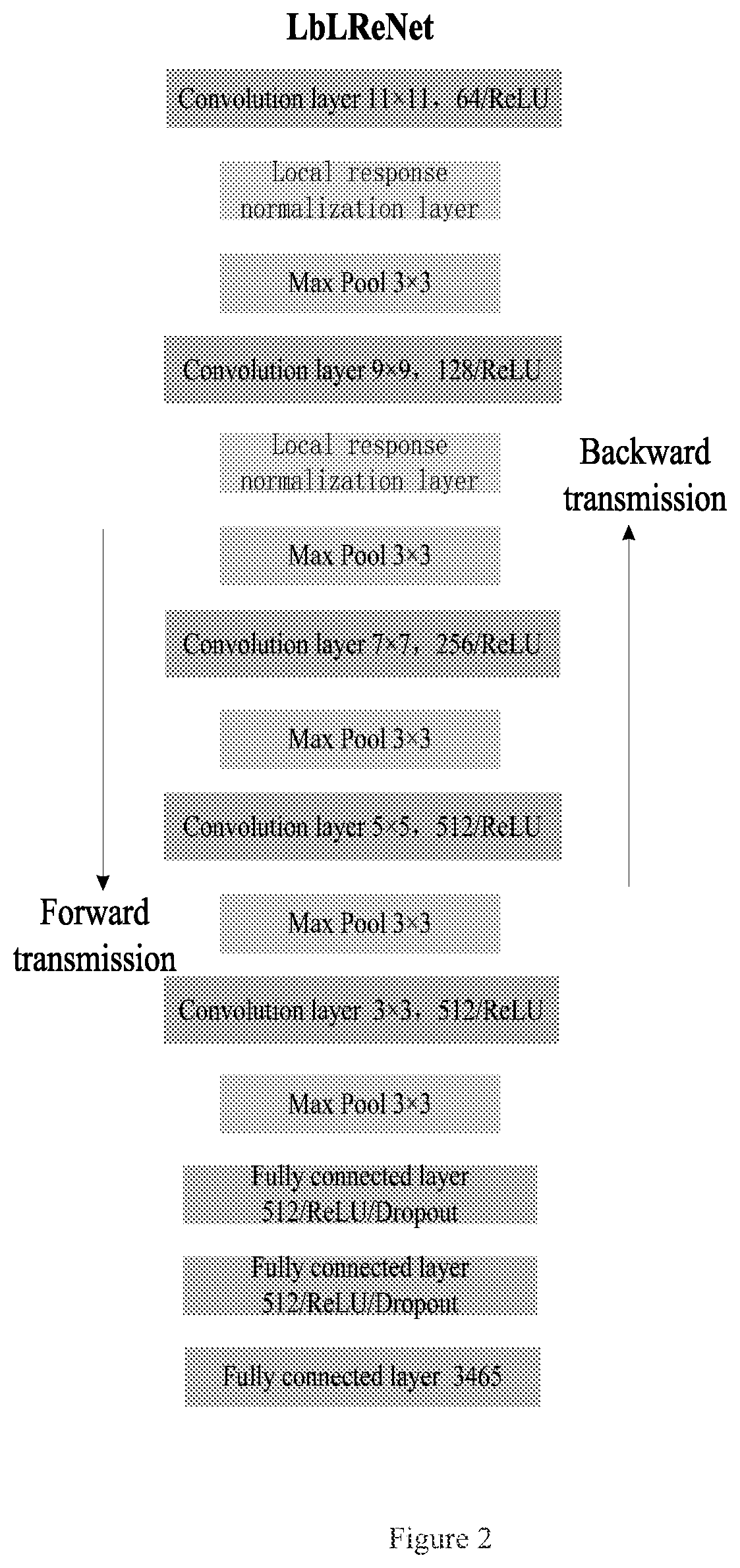
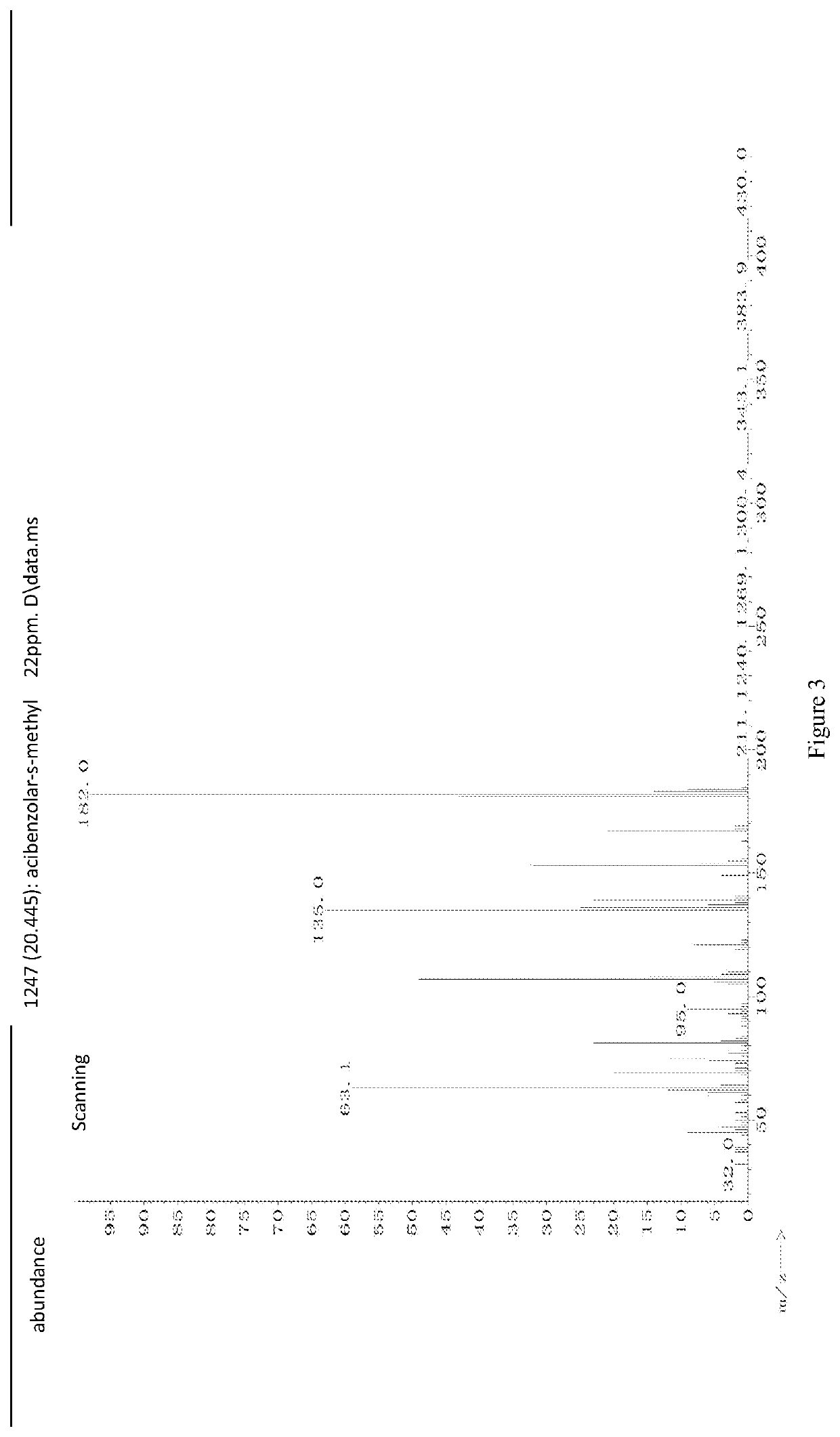
A cloud-platform based automatic identification system and method of seven types of mass spectrums for pesticides and chemical pollutants commonly used in the world
ActiveUS20220050092A1Ensure accuracy and reliabilityInterfere with detectionMolecular entity identificationComponent separationInformation repositoryPesticide residue
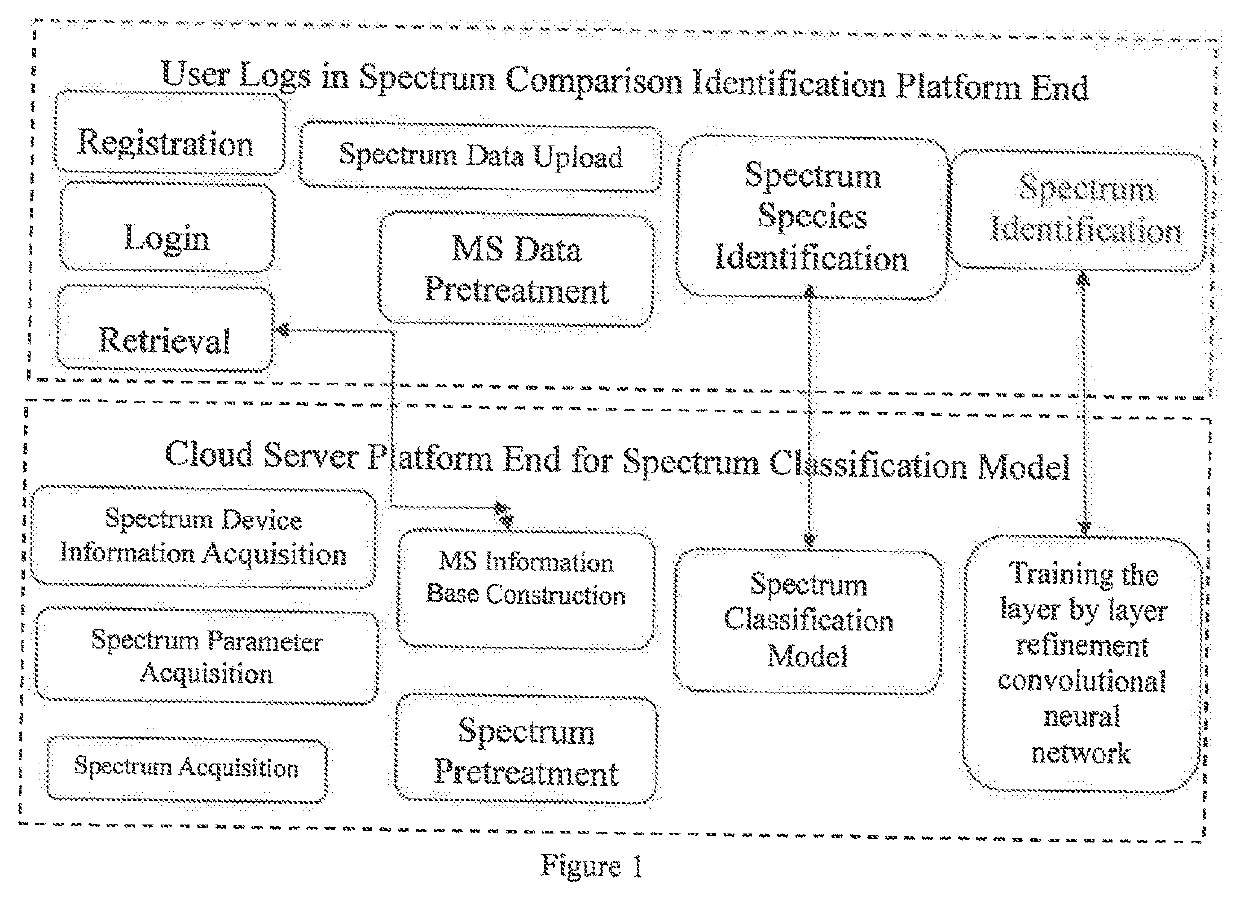
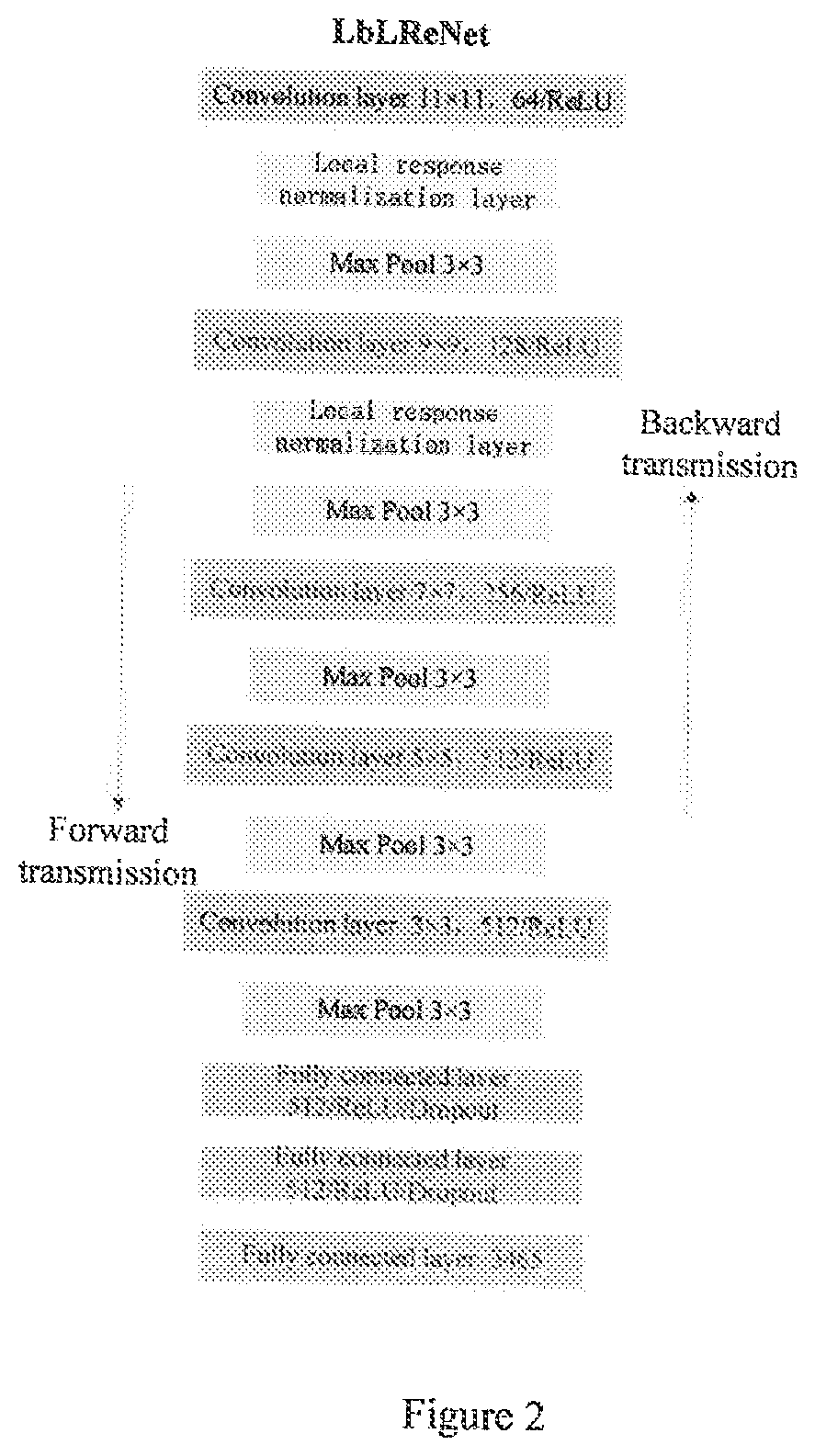
A cloud server platform end is used to construct a mass spectrum species classification model, extract a mass spectrum data feature, and construct a training model of the convolutional neural network; a user platform end is used to upload the mass spectrum, experiment condition and device data, directly screen and identify the type of the mass spectrum based on the mass spectrum species classification model or the mass spectrum information base, automatically compare and identify the species and name of the pesticides based on the neural network model trained by the cloud server platform end, and feedback the comparison result to the user. The disclosure solves the restriction on the purchase of standards for user, the use of the system is not limited by the location, and the pesticide residues could be detected automatically, quickly and accurately.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE +1
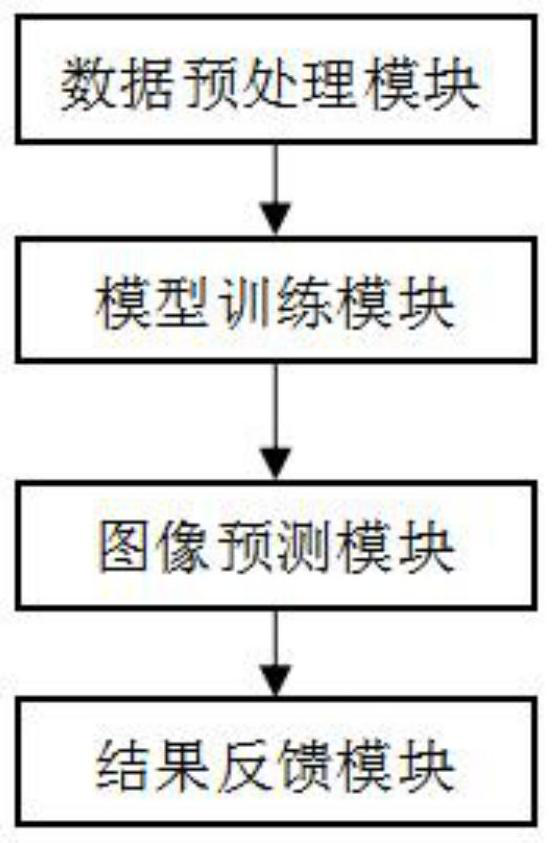
Hierarchical structure-based two-stage forestry pest identification and detection system and method
InactiveCN113449806AHigh feasibilityImprove recognition accuracyCharacter and pattern recognitionNeural architecturesForest industryData set
The invention discloses a hierarchical structure-based two-stage forestry pest identification and detection system, and the system comprises: a data preprocessing module which is used for preprocessing a labeled forestry pest image data set, and obtaining a first data set for training a target detection model in a pest detection stage and a second data set for training a classification model in a pest classification stage; the model training module that is used for training a target detection model in a pest detection stage by utilizing the first data set and training a classification model in a pest classification stage by utilizing the second data set; and the image prediction module that is used for performing target detection on the test image by using the trained target detection model, extracting the insect part from the test image to obtain a new image, and performing species classification on the new image by using the trained classification model. The hierarchical structure -based two-stage forestry pest identification and detection system is high in feasibility, can greatly improve the recognition accuracy of pests, and has expansibility.
Owner:SUZHOU UNIV
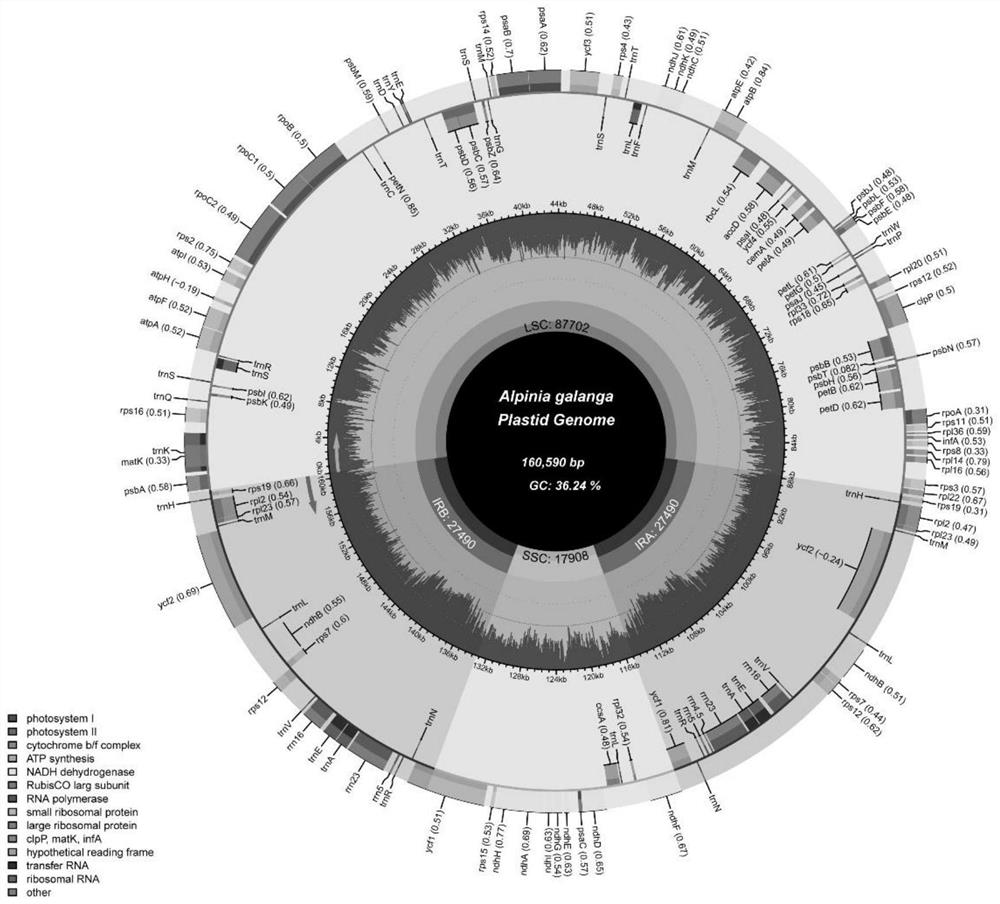
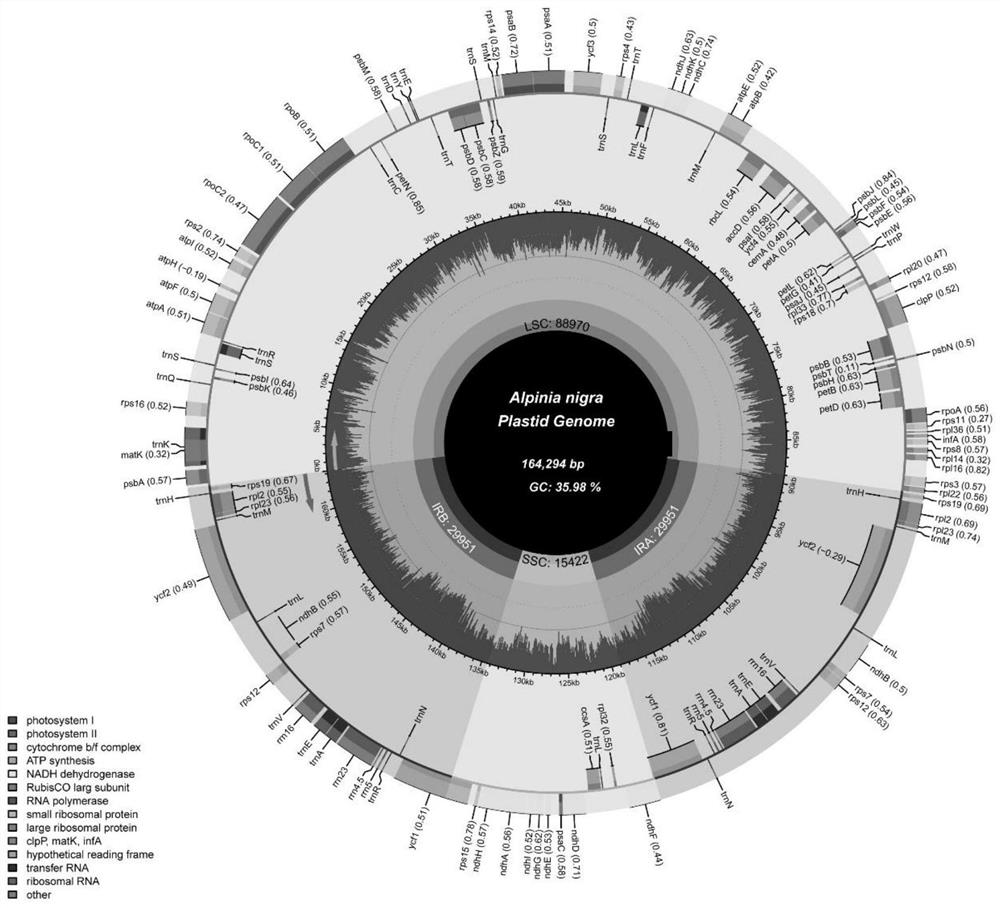
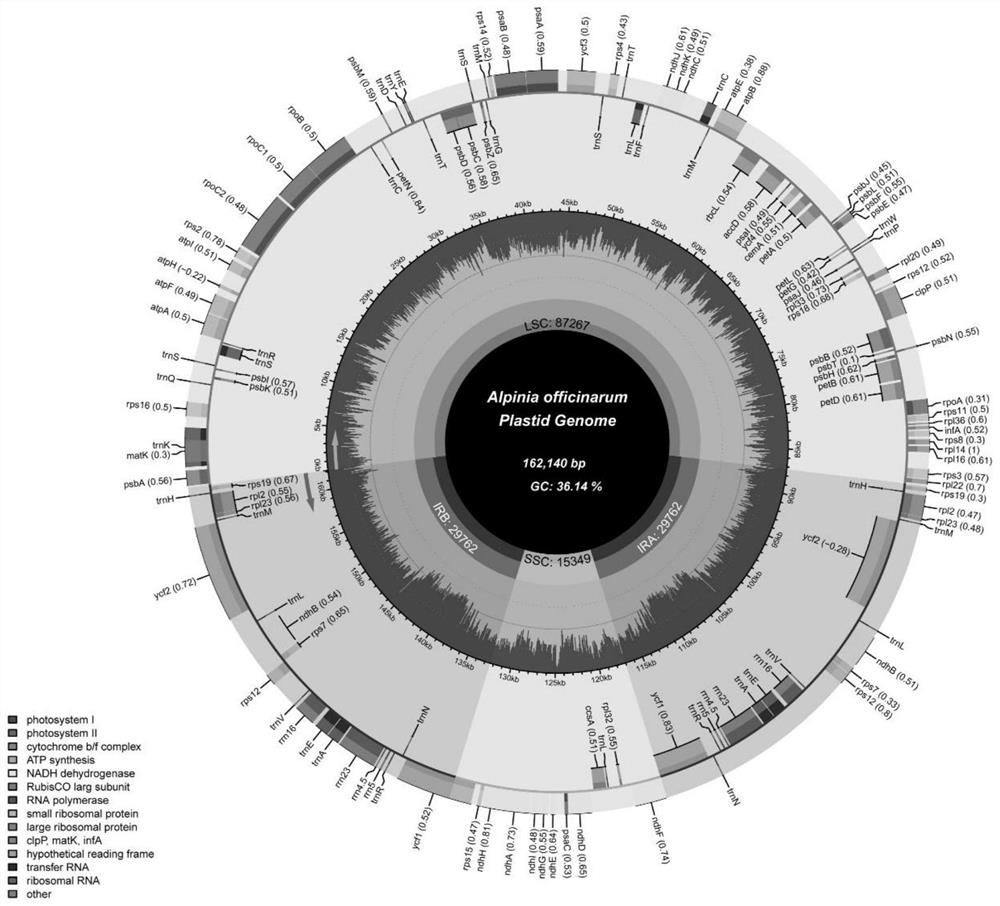
Molecular markers for identifying five alpinia plants and application thereof
ActiveCN113136451AEasy to classifyImprove discriminationMicrobiological testing/measurementAgainst vector-borne diseasesBiotechnologyMolecular identification
The invention discloses molecular markers for identifying five alpinia plants and application of the molecular markers, and belongs to the technical field of plant molecular identification. The molecular markers and primers designed based on the molecular markers can effectively identify and distinguish fructus galangae, alpinia hainanensis, alpinia black fruit, galangal and alpinia oxyphylla, and are beneficial to alpinia species classification, plastid genome evolution research and medicinal product discrimination derived from alpinia species.
Owner:INST OF MEDICINAL PLANT DEV CHINESE ACADEMY OF MEDICAL SCI
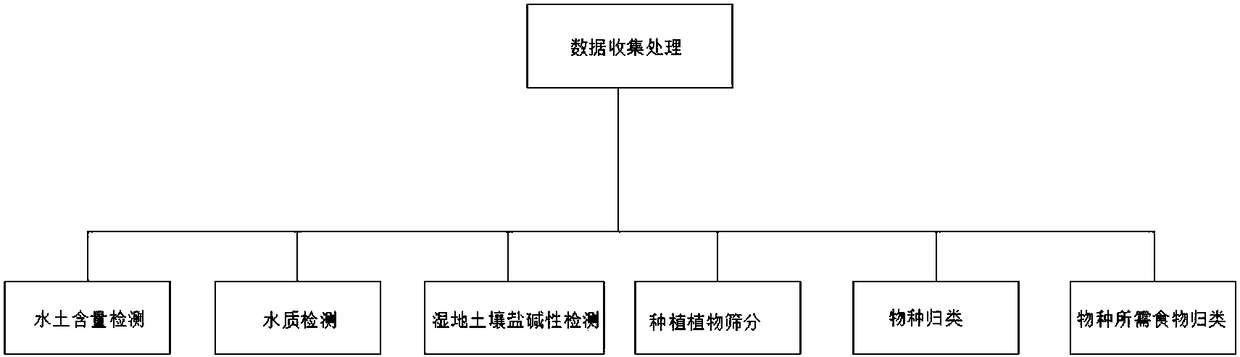
Wetland ecosystem restoration method by soil seed banks
ActiveCN108496702AImprove survival rateAvoid water and soil erosion, etc.Climate change adaptationPlant cultivationSpecies classificationEcosystem
The invention discloses a wetland ecosystem restoration method by soil seed banks, and relates to the technical field of soil seed banks. The wetland ecosystem restoration method by the soil seed banks comprises soil water content detection, water quality detection, wetland soil salt alkaline detection, screening of planted plants, species classification, and classification of food required by species. A certain water flow and soil are needed for the growth of plants, so that when plants are needed to be planted in a wetland, soil water content detection is carried out on the soil in differentareas in the wetland. According to the wetland ecosystem restoration method by the soil seed banks, in order to ensure the sustainable development of the wetland ecosystem and the survival rate of soil seeds, comprehensive detection and monitoring are carried out on water quality in the wetland, moisture content in soil, water-soil isolation, habitat species classification, species eaten by the habitat species, and the salt-alkaline content in the soil, so that the survival rate of the soil seed banks is increased by 20%, and the situations such as water and soil loss and the like in the wetland are avoided.
Owner:HUNAN ACAD OF FORESTRY
Classified conservation device for machine parts
InactiveCN107717925AIncreased load-bearing capacityExtended service lifeWork tools storageInterior spaceMachine parts
The invention relates to the technical field of mechanical accessories storage devices, in particular to a sorted storage device for mechanical accessories. A rope buckle is fixedly installed, a handle is fixedly installed on the outer surface of the front end of the main body of the device, clamps are fixedly installed on both sides of the outer surface of the front end of the main body of the device, and a buckle is movably installed on the outer surface of the upper end of the clamp. The interior of the main body of the device is fixedly equipped with a thickened plate. A sorted storage device for mechanical accessories according to the present invention is provided with a thickened plate, a classified nameplate and a folding baffle, which can increase the load-bearing strength and service life of the device, and make the The staff can intuitively understand the information of various mechanical accessories, and can also easily change the size of the internal space, which is suitable for different working conditions and brings better prospects for use.
Owner:无锡市天密石化通用件厂
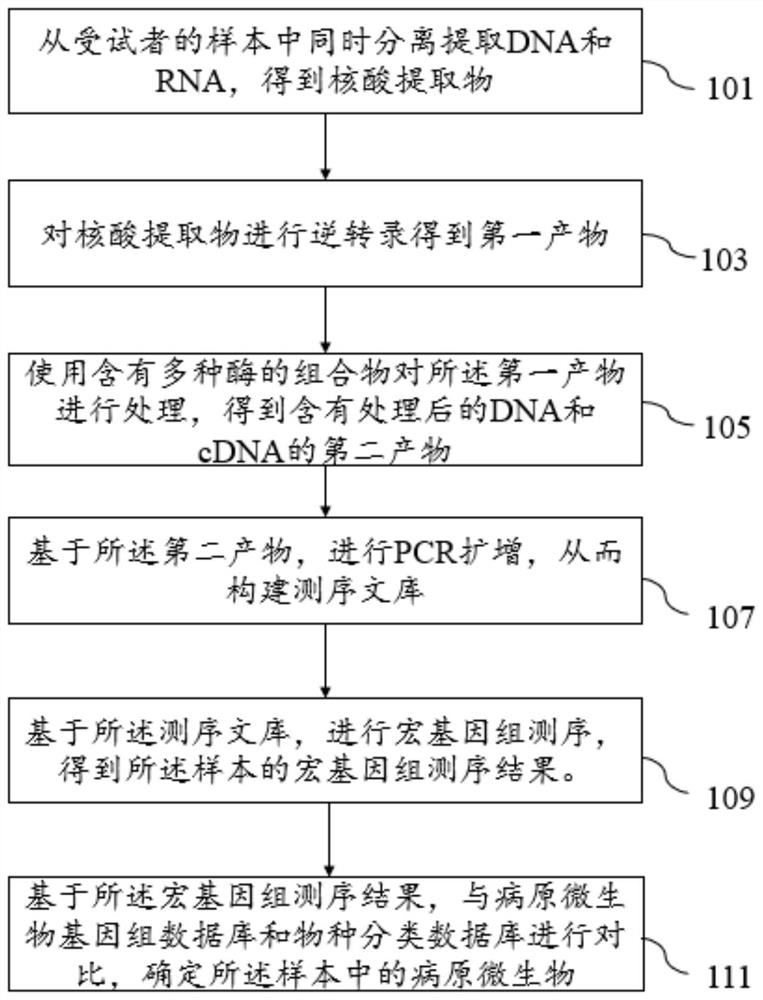
Method for high-throughput rapid detection of pathogenic microorganisms
InactiveCN113215235AMicrobiological testing/measurementProteomicsGenomic sequencingT4 polynucleotide kinase
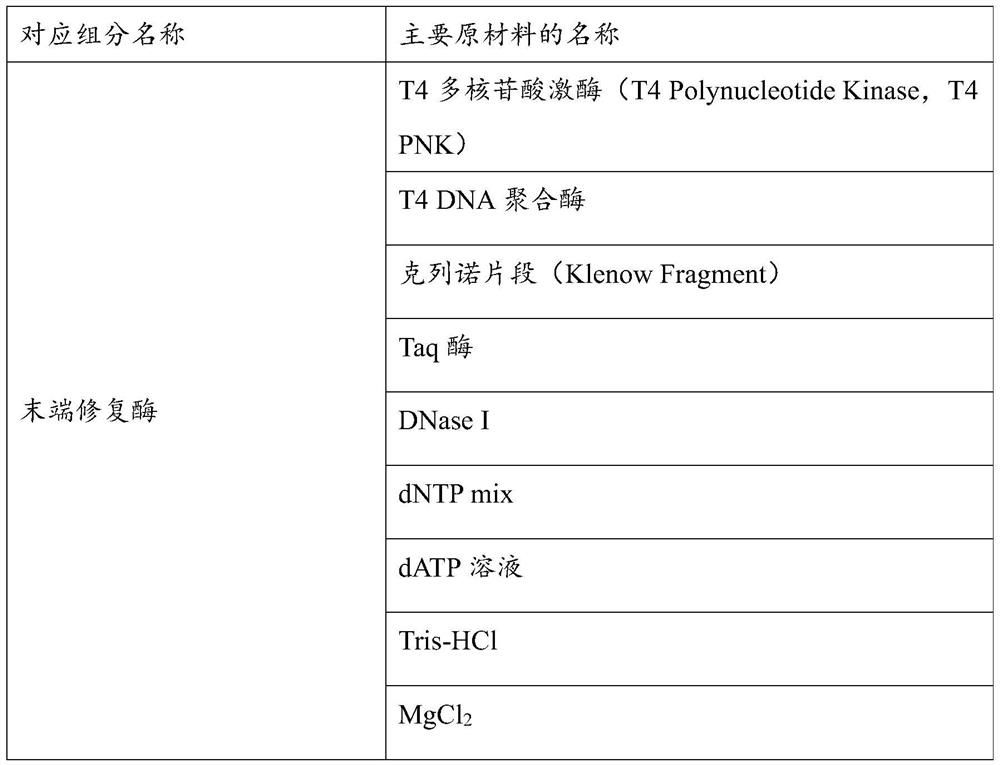
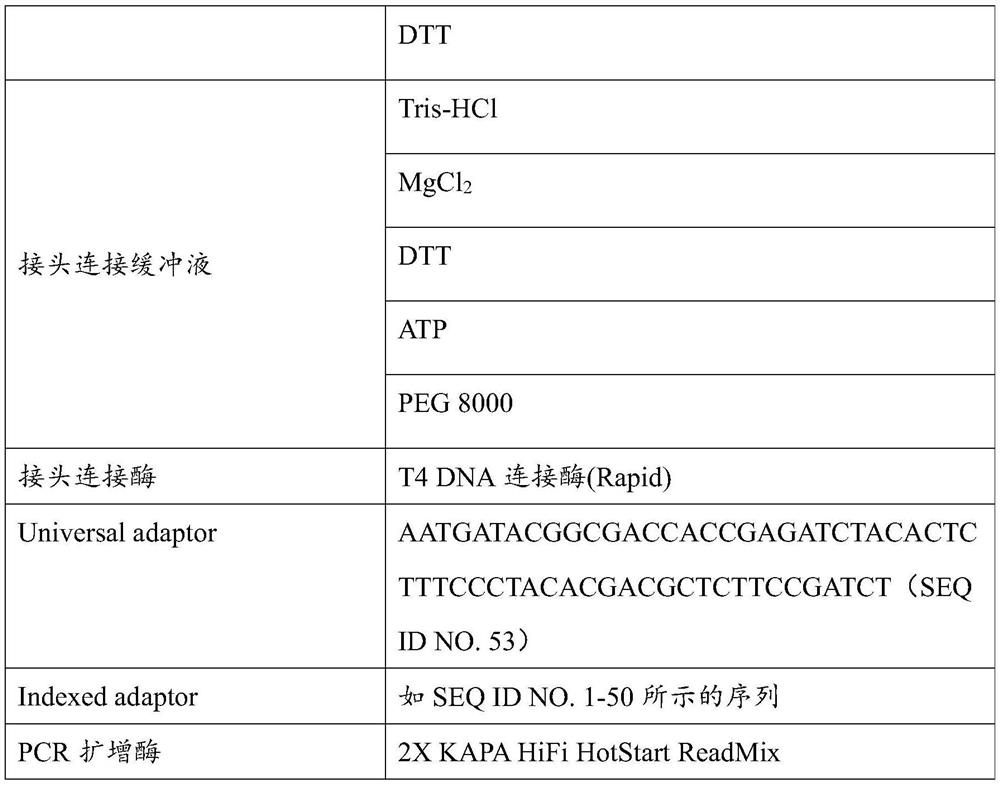
The invention discloses a method for detecting pathogenic microorganisms. The method comprises the following steps: simultaneously separating and extracting DNA and RNA from a sample of a subject; performing reverse transcription on the extracted RNA to obtain cDNA; treating the extracted DNA and the extracted cDNA by using a composition containing a plurality of enzymes, so as to obtain treated DNA and treated cDNA; performing PCR amplification on the basis of the treated DNA and the treated cDNA so as to construct a sequencing library; performing metagenome sequencing on the basis of the sequencing library to obtain a metagenome sequencing result of the sample; and determining the pathogenic microorganisms in the sample based on the metagenome sequencing result, a pathogenic microorganism genome database and a species classification database. The invention also discloses a kit for establishing the microbial metagenome sequencing library. The kit comprises a fragmentation and terminal repair system, wherein the fragmentation and terminal repair system comprises DNA incision enzymes, T4 polynucleotide kinase, T4 DNA polymerase, a Klinno fragment and Taq enzymes.
Owner:JIAXING YUNYING MEDICAL INSPECTION CO LTD
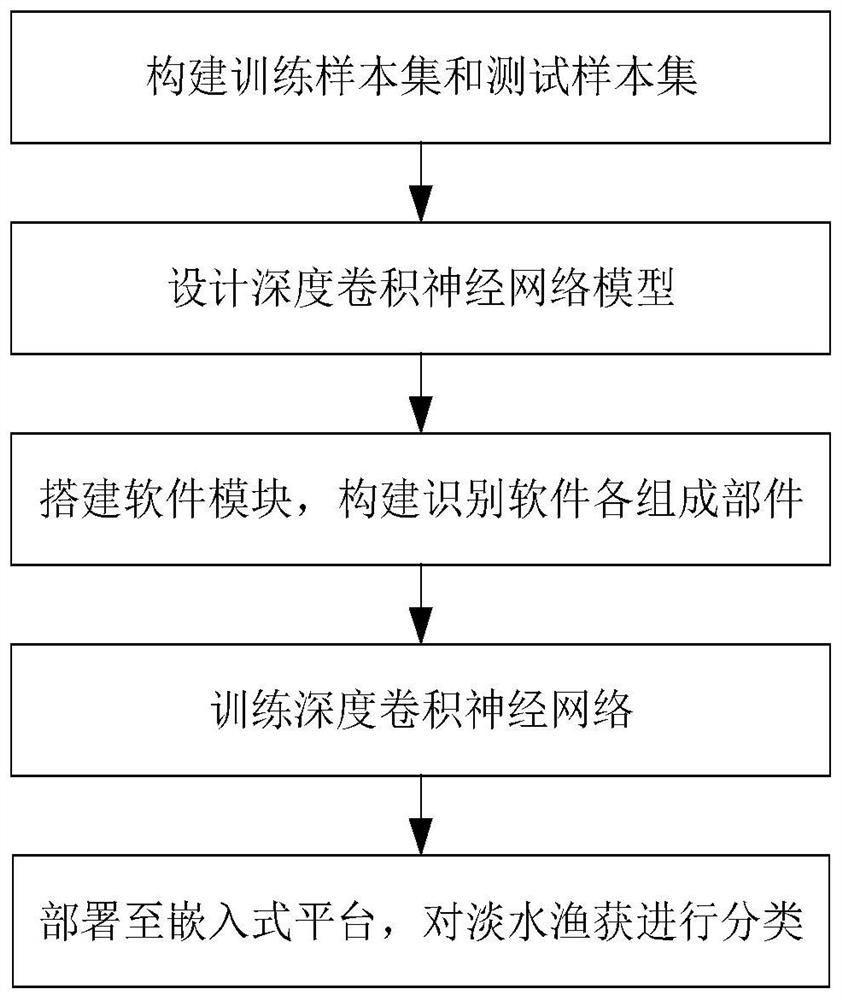
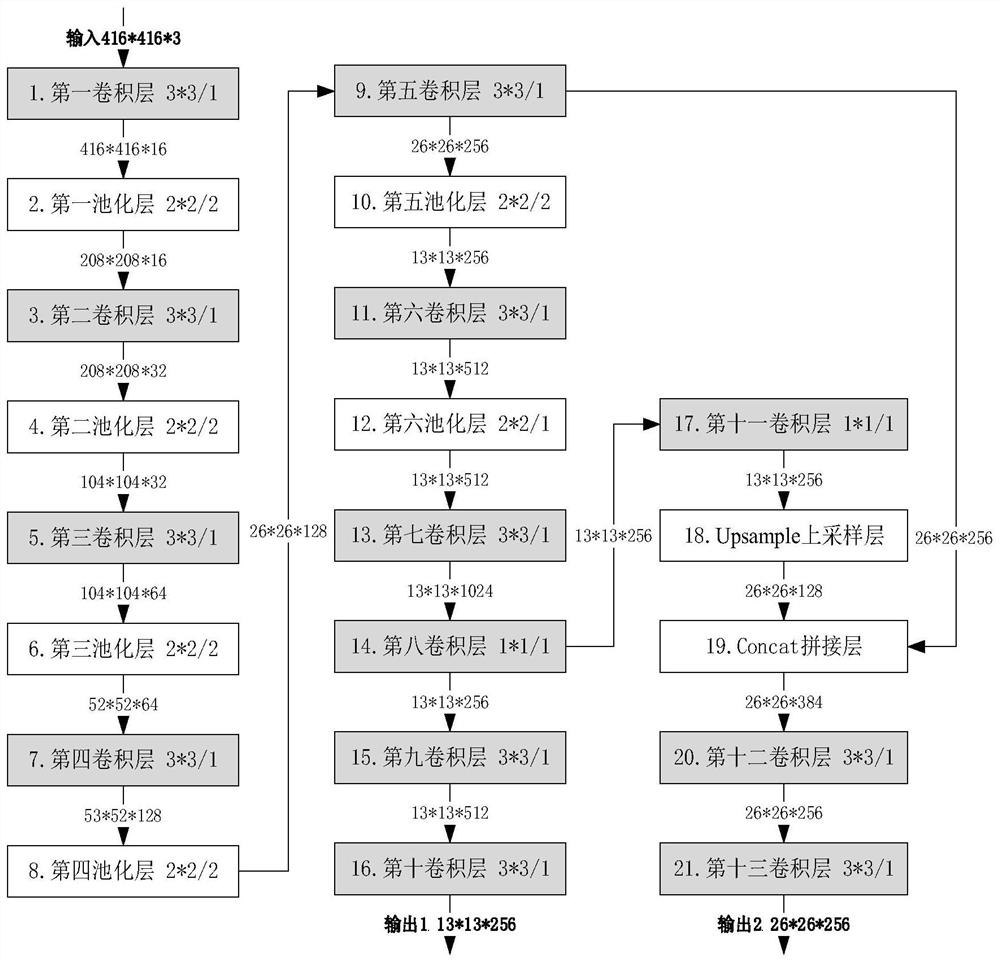
Freshwater fish image real-time identification method based on lightweight convolutional network
PendingCN112966698AAvoid missingOvercome unusableCharacter and pattern recognitionNeural architecturesData setFeature extraction
The invention discloses a freshwater fish image real-time identification method based on a lightweight convolutional network, and aims to solve the problems of lack of data sets, difficulty in feature extraction and low identification speed in the existing freshwater fish species classification technology. The method comprises the following specific steps: (1) constructing a freshwater fish image data set; (2) constructing a lightweight deep convolutional neural network; (3) training a deep convolutional neural network; (4) identifying the freshwater fish test image in real time; by constructing the freshwater fish image data set and utilizing the trained lightweight deep convolutional neural network, real-time recognition is automatically performed on the collected freshwater fish image, and the method has the advantages of no need of manually extracting fish body features, high recognition precision, high speed and small hardware resource consumption.
Owner:XIDIAN UNIV
Tailored artificial intelligence
A system and method determine a classification by simulating a human user. The system and method translate an input segment such as speech into an output segment such as text and represents the frequency of words and phrases in the textual segment as an input vector. The system and method process the input vector and generate a plurality of intents and a plurality of sub-entities. The processing of multiple intents and sub-entities generates a second multiple of intents and sub-entities that represent a species classification. The system and method select an instance of an evolutionary model as a result of the recognition of one or more predefined semantically relevant words and phrases detected in the input vector.
Owner:PROGRESSIVE CASUALTY INSURANCE
Cloud-platform based automatic identification system and method of seven types of mass spectrums for pesticides and chemical pollutants commonly used in the world
ActiveUS11340201B2Ensure accuracy and reliabilityInterfere with detectionMolecular entity identificationComponent separationInformation repositoryPesticide residue
A cloud server platform end is used to construct a mass spectrum species classification model, extract a mass spectrum data feature, and construct a training model of the convolutional neural network; a user platform end is used to upload the mass spectrum, experiment condition and device data, directly screen and identify the type of the mass spectrum based on the mass spectrum species classification model or the mass spectrum information base, automatically compare and identify the species and name of the pesticides based on the neural network model trained by the cloud server platform end, and feedback the comparison result to the user. The disclosure solves the restriction on the purchase of standards for user, the use of the system is not limited by the location, and the pesticide residues could be detected automatically, quickly and accurately.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com