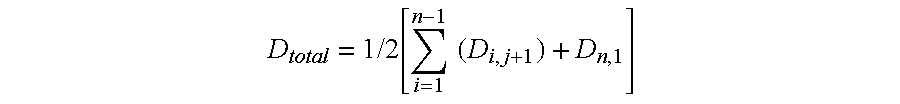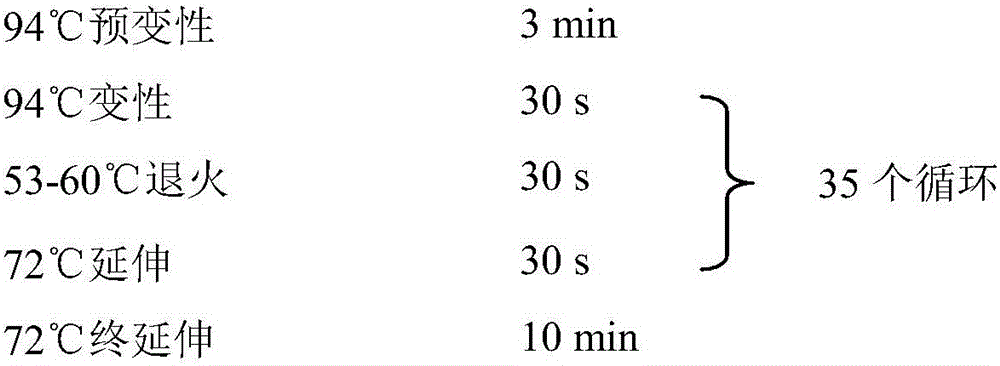Patents
Literature
54 results about "Phylogenetic relationship" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Phylogenetic relationships are the relationships that show how far back two species shared a common ancestor.
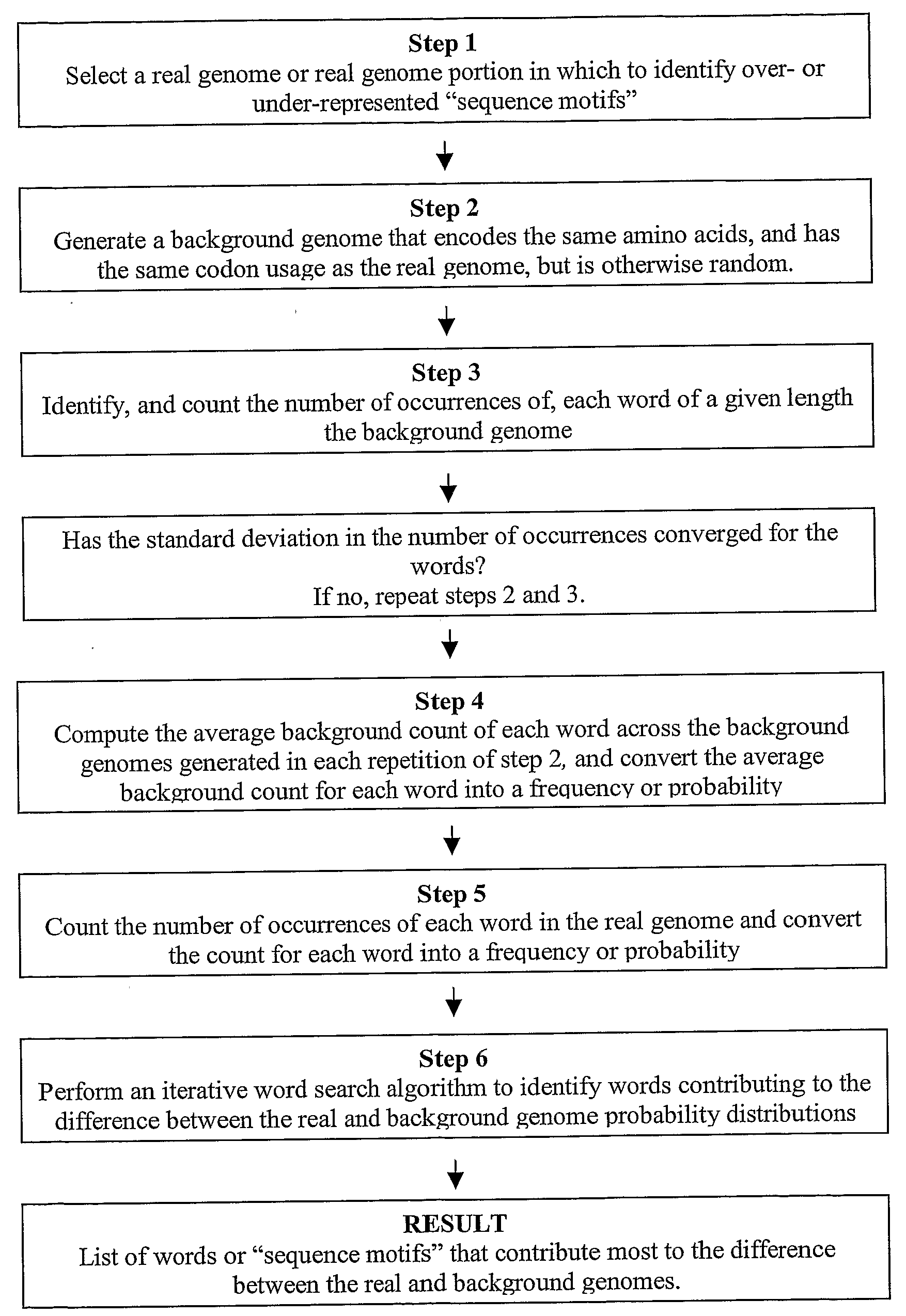
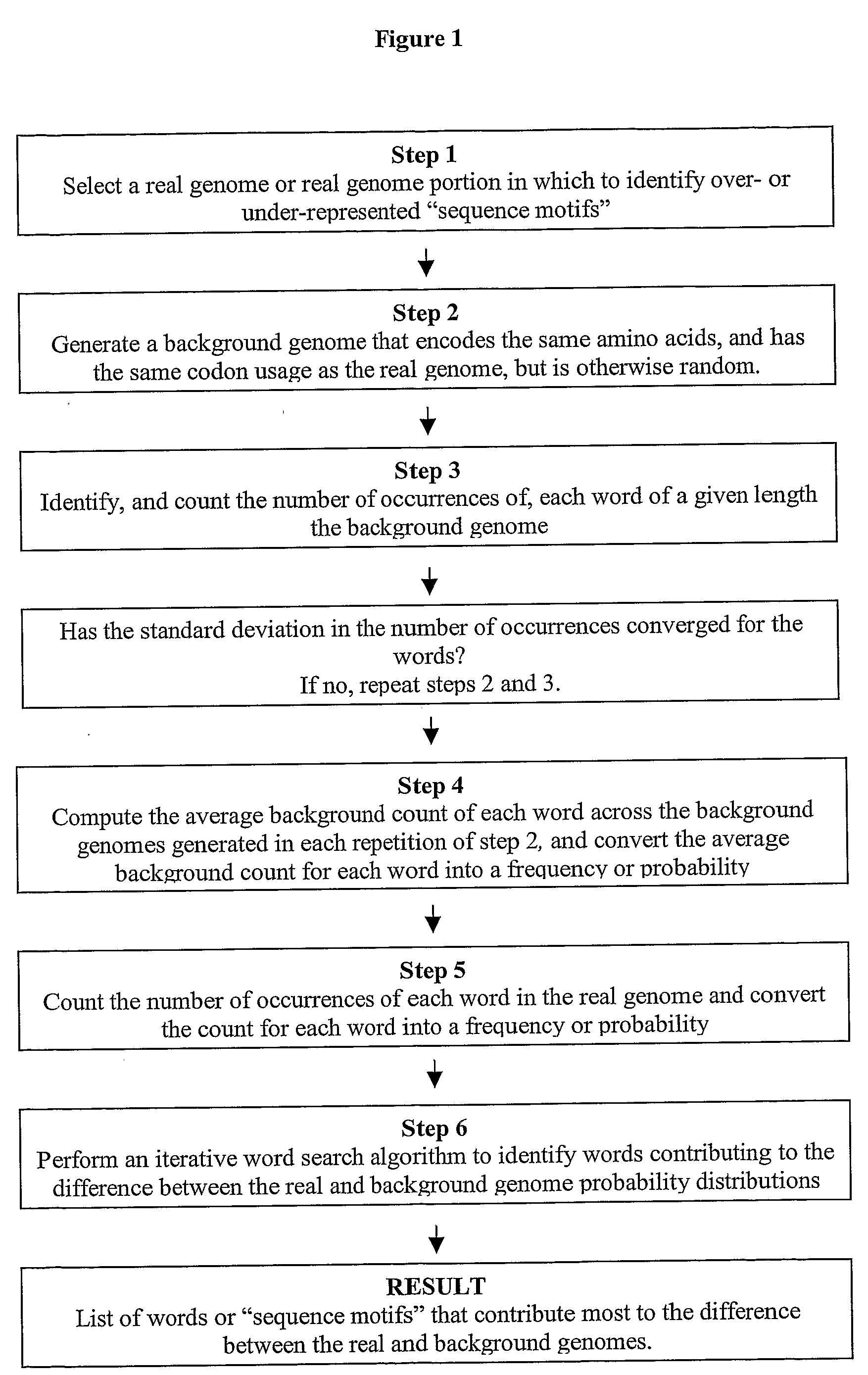
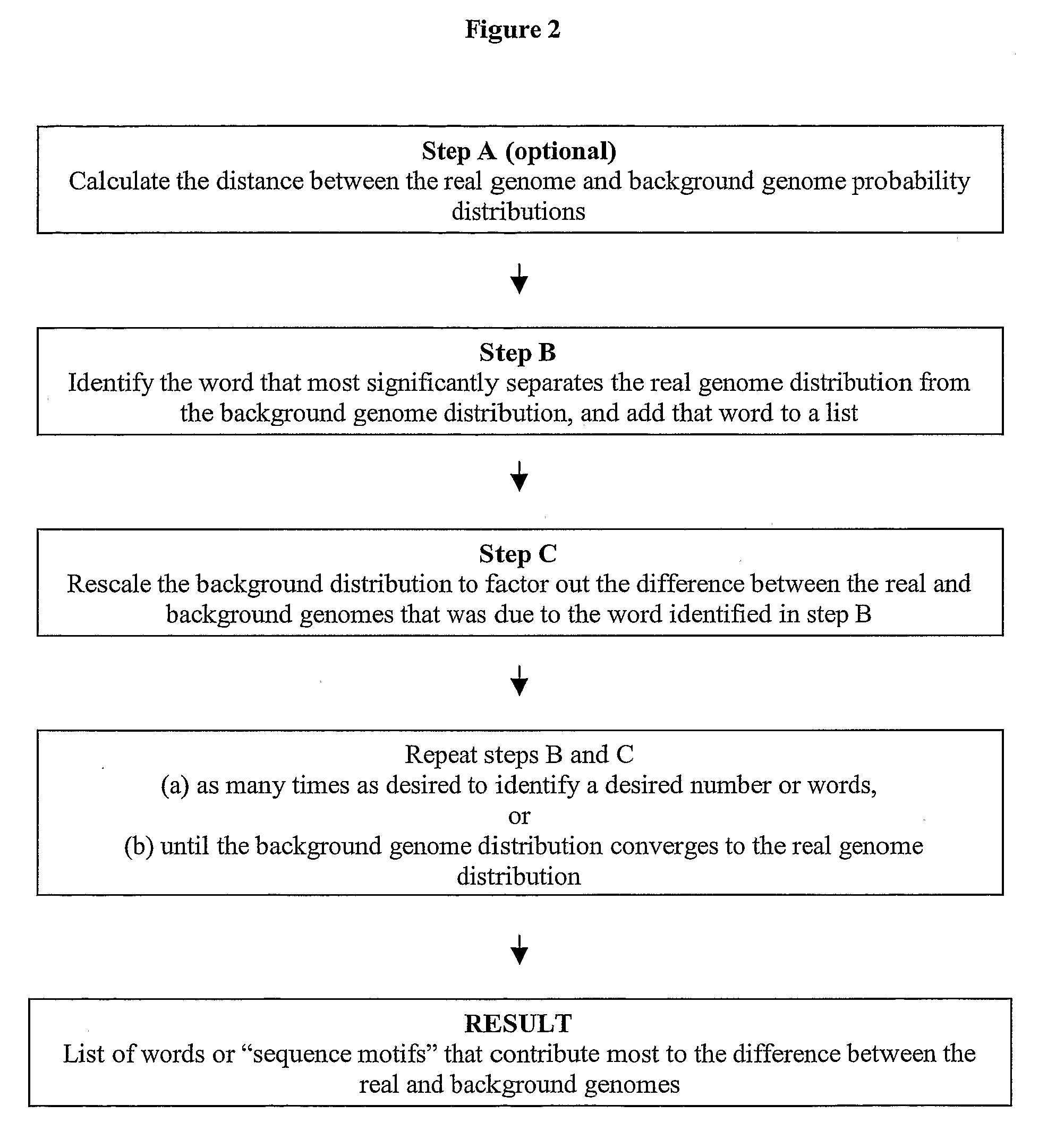
Methods for identifying sequence motifs, and applications thereof
InactiveUS20090208955A1Reduce in quantityIncrease the number ofMicrobiological testing/measurementProteomicsBinding siteInstability
The present invention relates to methods and algorithms that can be used to identify sequence motifs that are either under- or over-represented in a given nucleotide sequence as compared to the frequency of those sequences that would be expected to occur by chance, or that are either under- or over-represented as compared to the frequency of those sequences that occur in other nucleotide sequences, and to methods of scoring sequences based on the occurrence of these sequence motifs. Such sequence motifs may be biologically significant, for example they may constitute transcription factor binding sites, mRNA stability / instability signals, epigenetic signals, and the like. The methods of the invention can also be used, inter alia, to classify sequences or organisms in terms of their phylogenetic relationships, or to identify the likely host of a pathogenic organism. The methods of the present invention can also be used to optimize expression of proteins.
Owner:INST FOR ADVANCED STUDY
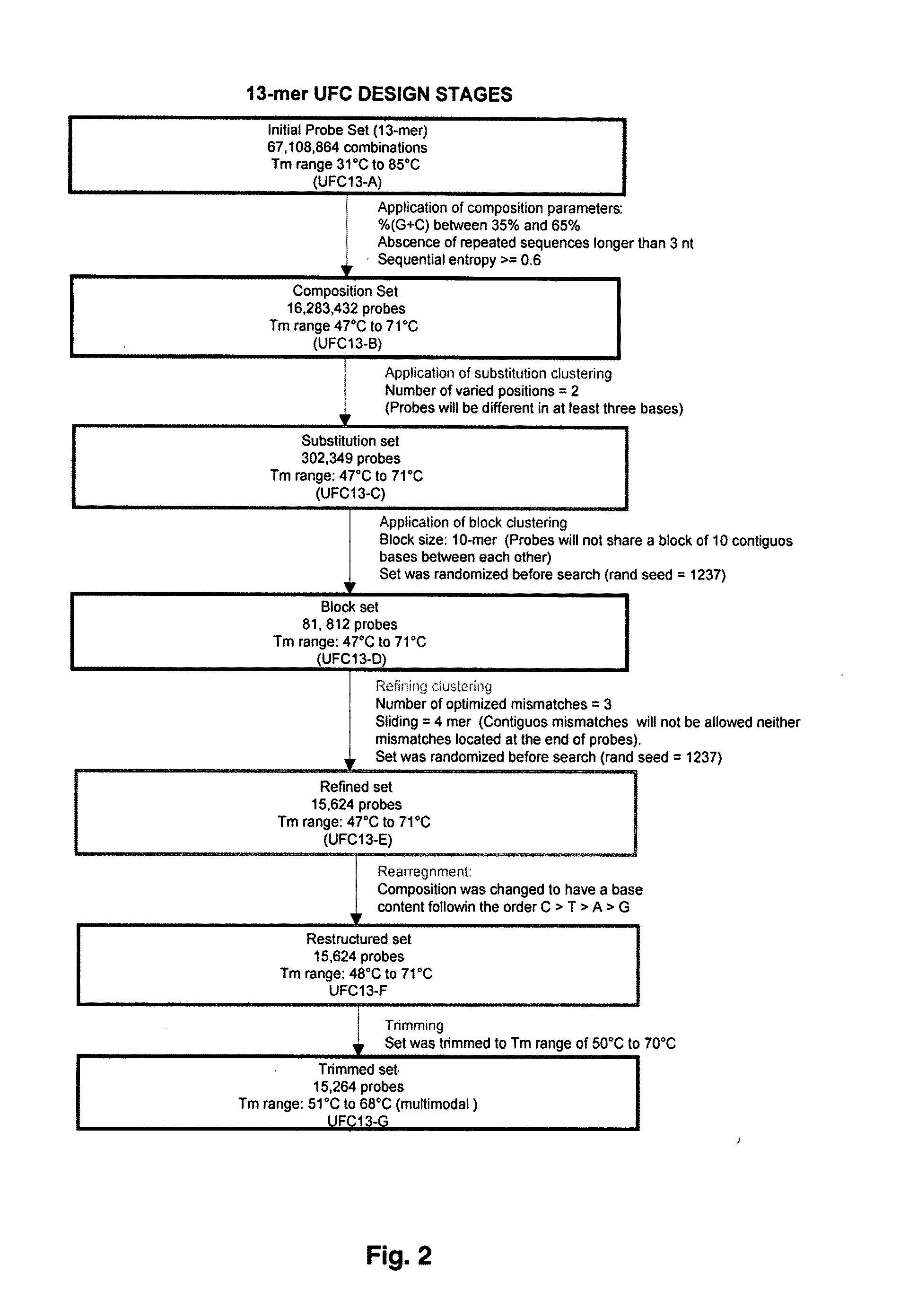
Universal fingerprinting chips and uses thereof
InactiveUS20110105346A1Avoid formingNucleotide librariesMicrobiological testing/measurementNucleotideOrganism
The present invention discloses a designing strategy for constructing a set of probes useful for analyzing all or most prokaryotic and eukaryotic genomes. A set of capture probes with optimal fingerprinting properties and highly representative of all possible sequences of an organism can be selected by six sequential steps. Fingerprinting potential of such probes is validated by phylogenetic analysis, which generates results that strongly correlate with phylogenetic trees produced by sequence alignment. The probes generated by the instant methods can be used for detecting an organism, for establishing phylogenetic relationships between different organisms, for detection of single nucleotide polymorphisms and a wide variety of other applications that require genetic analysis.
Owner:BEATTIE KENNETH L +4
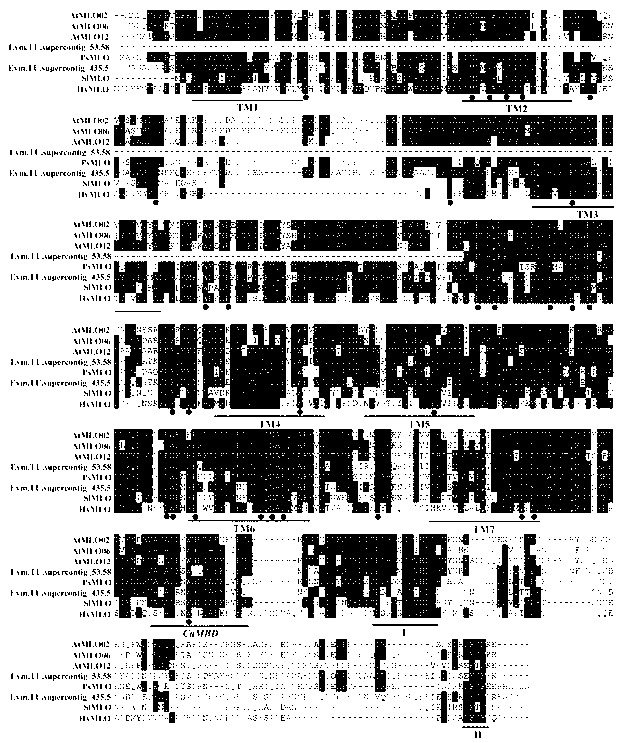
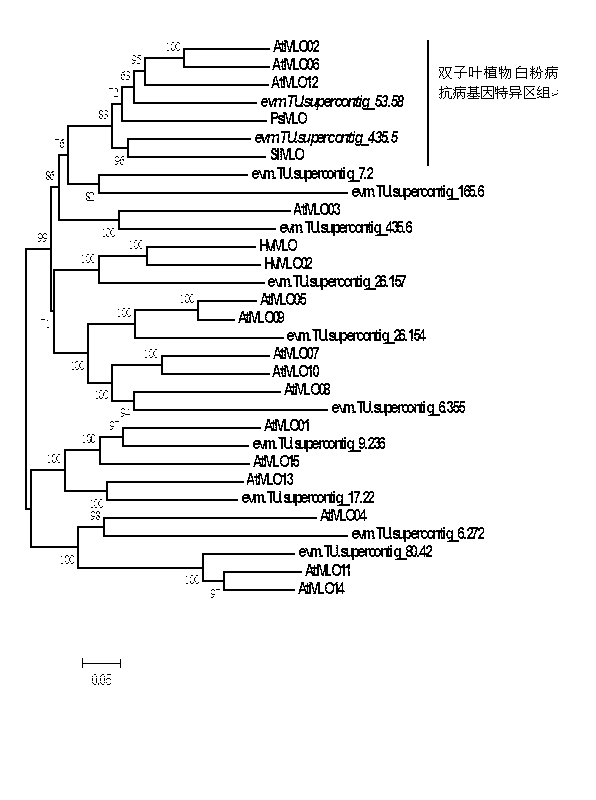
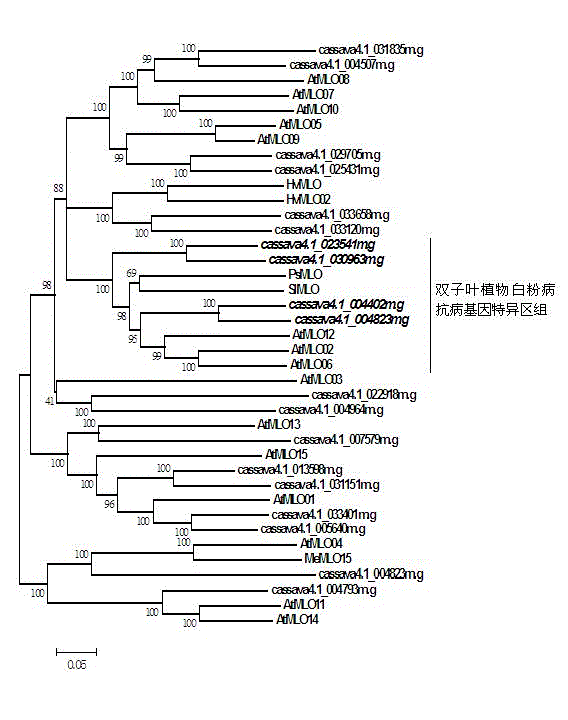
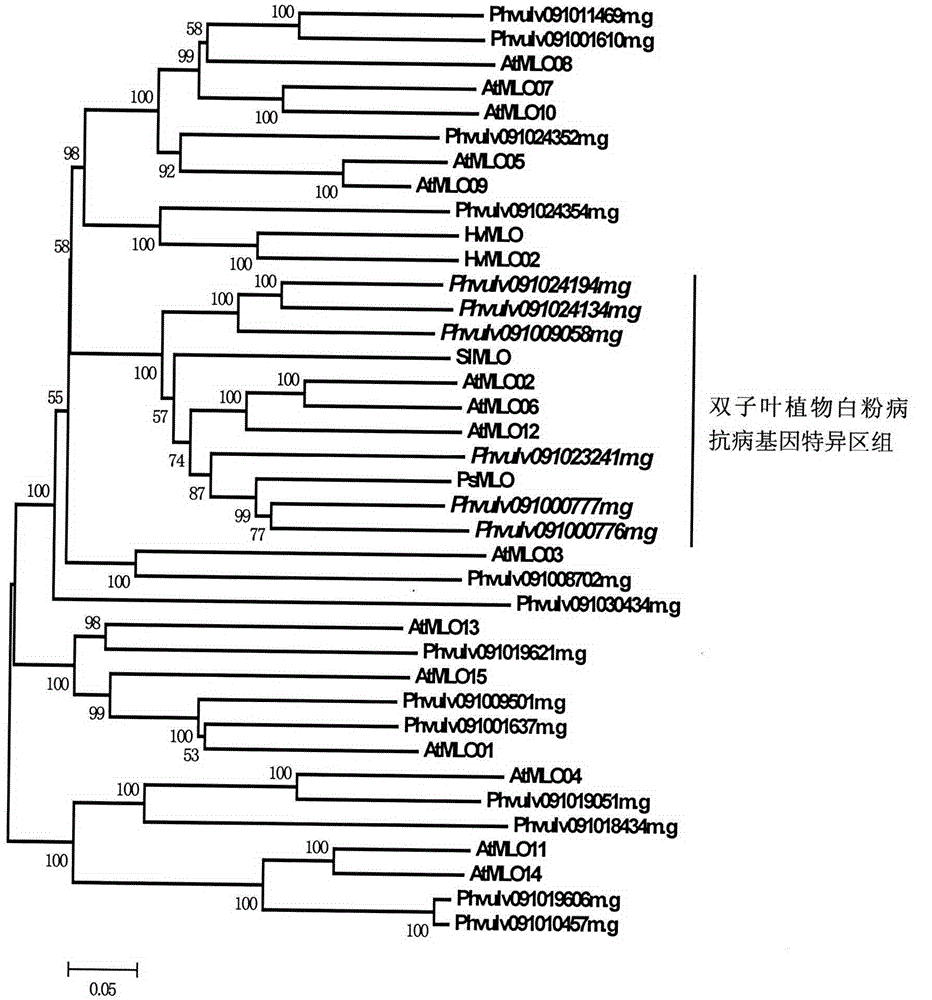
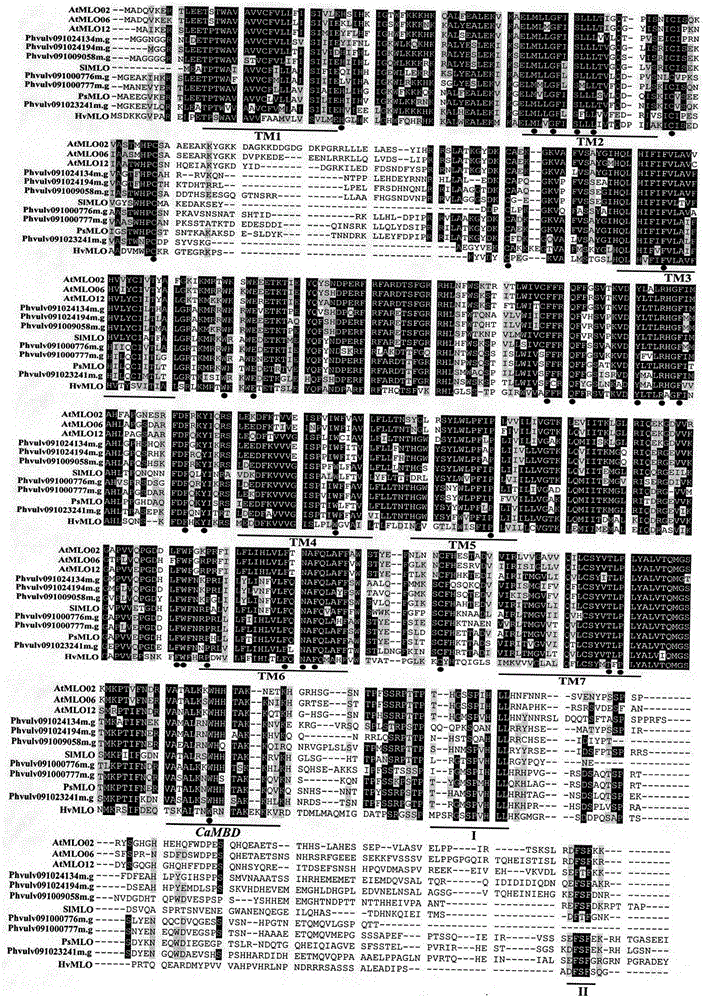
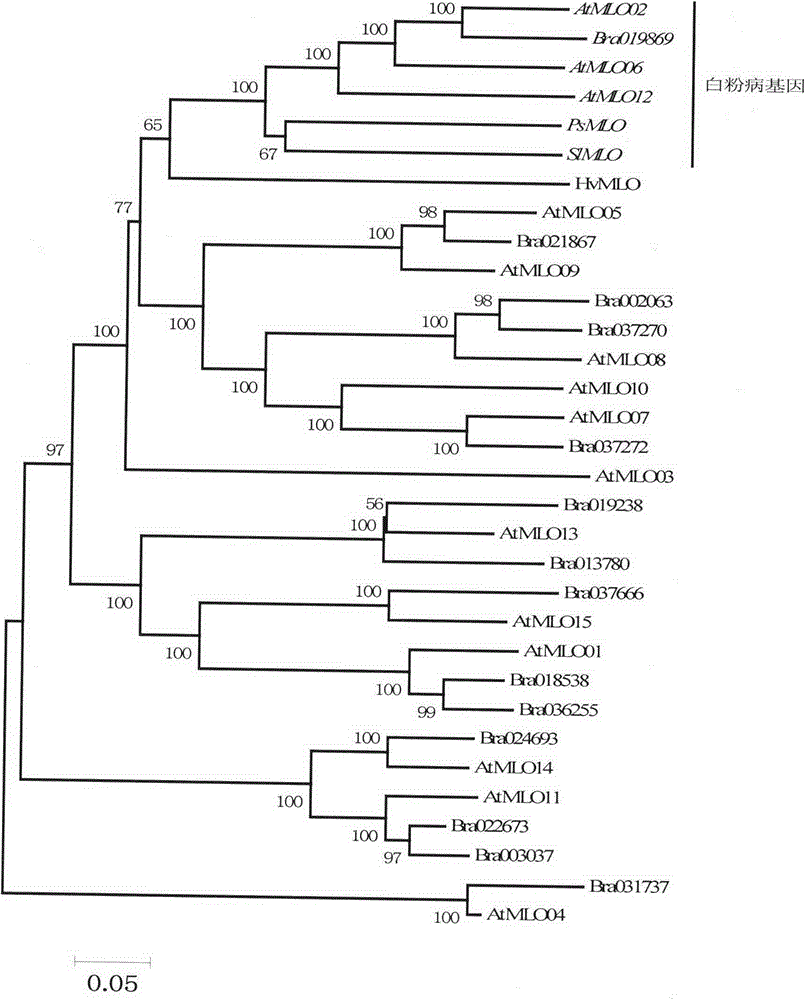
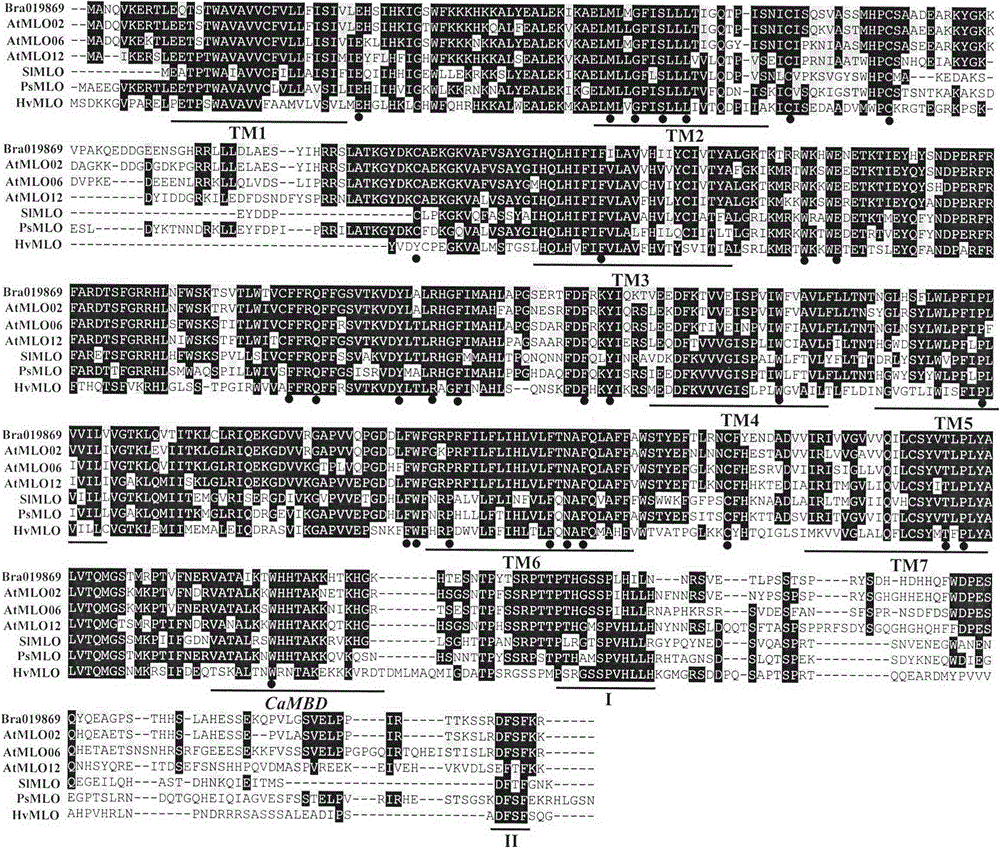
Application of bioinformatics in fast identifying powdery mildew resistance gene of carica papaya l
InactiveCN102719448AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesBiotechnologyPapaya family
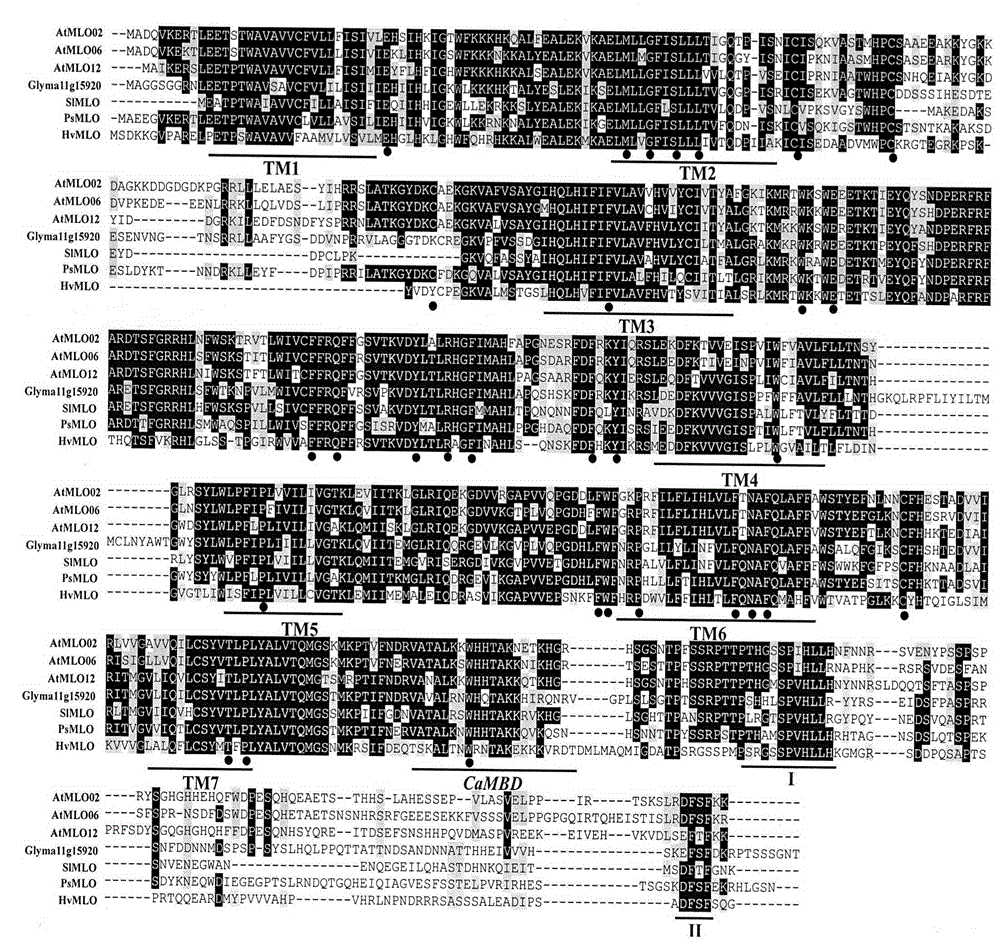
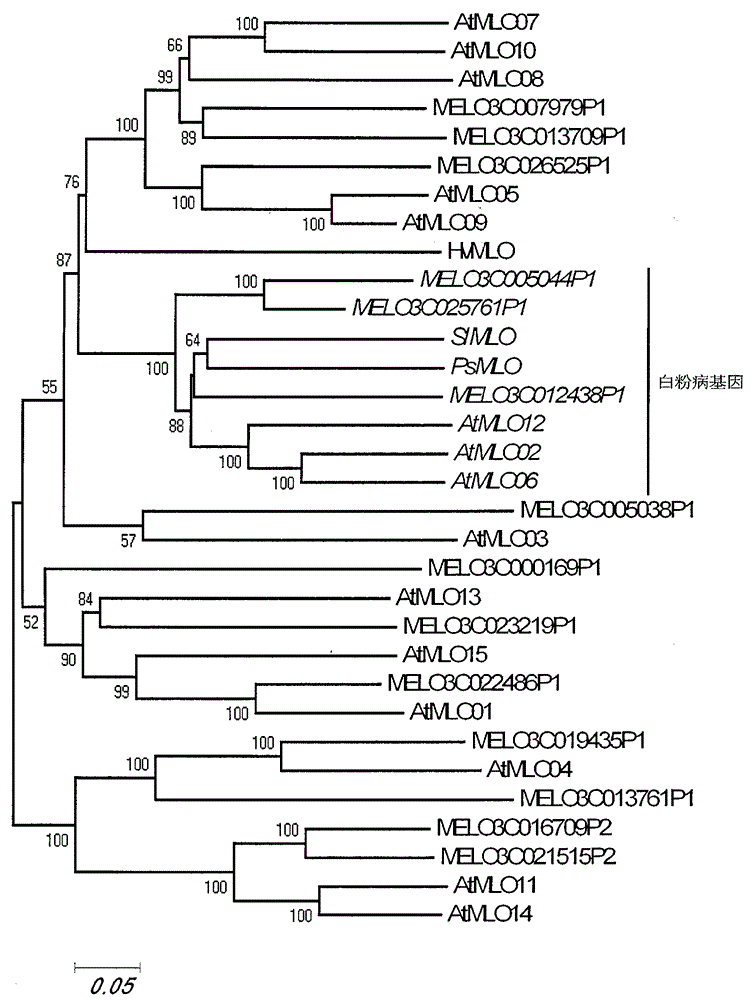
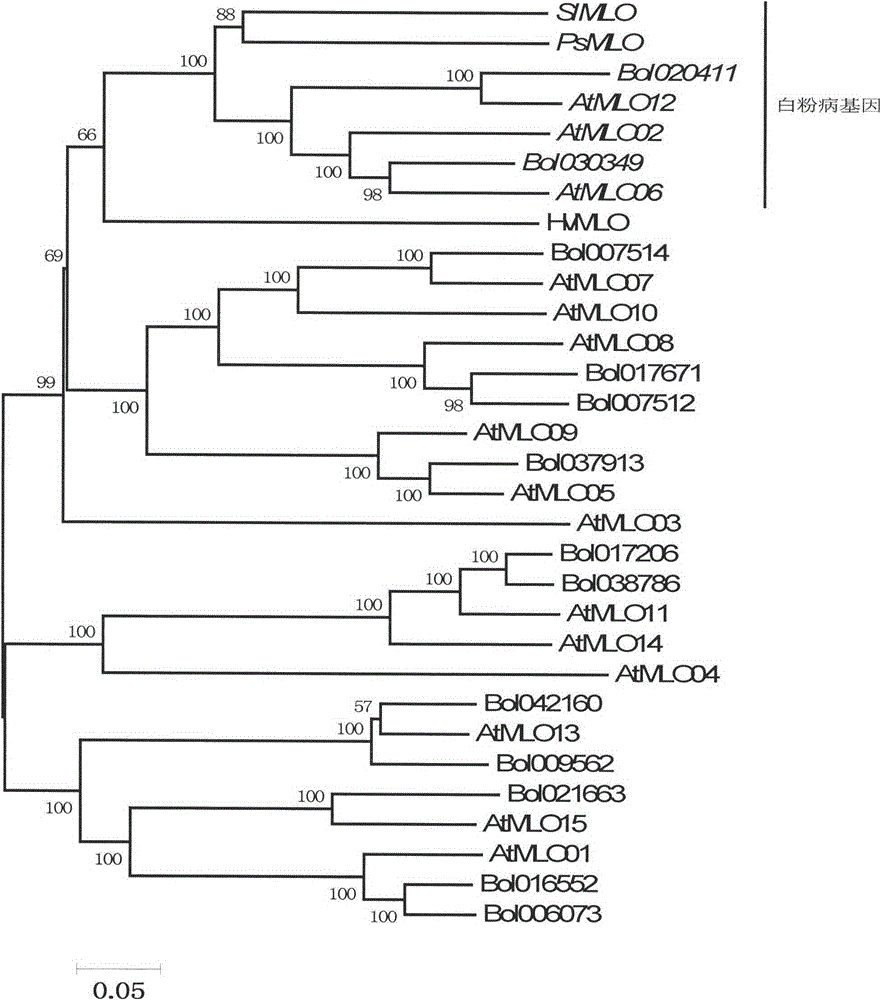
The invention relates to fast identification of a powdery mildew resistance gene of carica papaya l, relating to subject knowledge including plant comparative genomics, genetics, bioinformatics and the like, belonging to the field of plant biotechnology sciences. Application of the invention mainly comprises the steps: 1) downloading complete genome sequence of the carica papaya l and acquiring MLO-type genes; 2) identifying the MLO-type genes; 3) researching phylogenetic relationships of the MLO-type genes; and 4) comparing MLO-type powdery mildew resistance genes. The invention has the advantages that discovery period of the powdery mildew resistance gene of the carica papaya l is effectively shortened, which assists in fast identification of the powdery mildew resistance gene; that identified candidate powdery mildew resistance genes are developed with corresponding co-segregation functional markers (SNP, SCAR, etc.), which can be used in fast auxiliary selection of molecular markers of the powdery mildew resistance gene with high accuracy; that other molecular markers of resistance genes can be combined to formulate multi-resistance breeding materials, which shortens breeding years and improves breeding yields; and that a foundation is laid to illustrate the powdery mildew resistance gene of the carica papaya l.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
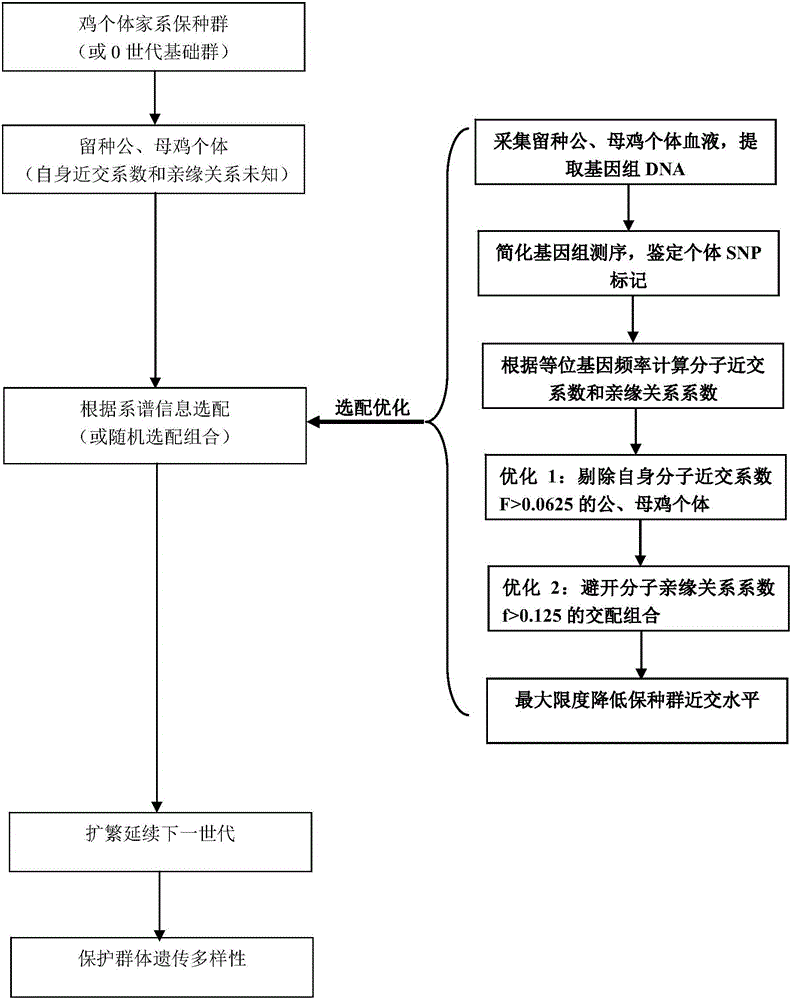
Chicken preserved population individual apolegamy optimizing method
ActiveCN106086172AReduce the level of inbreedingSolve the problem that individual fine matching cannot be carried outMicrobiological testing/measurementGenomic sequencingGenetic diversity
The invention discloses a chicken preserved population individual apolegamy optimizing method. Blood DNA of chicken preserved population breed reserving cock and hen individuals is extracted, sequencing is carried out through a reduced genomic sequencing technology, and the sequence is compared with a sequence of the GenBank database reference chicken breed to find SNP loci existing in different individuals. The individual own inbreeding coefficient and the interindividual phylogenetic relationship coefficient are calculated through gene frequency and genotype frequency of the SNP loci, individual apolegamy is optimized according to the statistics value, mating combination of individuals with an overlarge inbreeding coefficient and individuals with an overlarge within-family phylogenetic relationship coefficient is avoided, and the purposes of reducing the preserved population inbreeding level and maintaining the genetic diversity total quantity steady state are achieved. By means of the method, the problem that fine individual apolegamy cannot be carried out depending on genealogy information in the chicken breed resource protection process can be effectively solved, the preserved population inbreeding level can be effectively reduced, the genetic diversity steady state is maintained, and the breed preservation effect and efficiency are improved.
Owner:JIANGSU INST OF POULTRY SCI
Rapid identification of powdery mildew gene of Medicago truncatula by utilizing comparative genomics
InactiveCN102719445AQuick assist selectionImprove accuracyMicrobiological testing/measurementPlant peptidesBiotechnologyRapid identification
The present invention is rapid identification of a powdery mildew gene of Medicago truncatula, relates to discipline knowledge of plant comparative genomics, genetics and bioinformatics and belongs to the technical field of plant biotechnology. The invention mainly comprises the following steps: 1) download of Medicago truncatula genome sequence and collection of an MLO gene; 2) identification of MLO type gene; 3) MLO gene phylogenetic relationship; and 4) comparison of MLO type powdery mildew gene. The invention effectively shortens a mining cycle of the powdery mildew gene of Medicago truncatula and facilitates the rapid identification of the powdery mildew gene. The identified candidate powdery mildew genes can be used to develop corresponding coseparation functional markers (SNP and SCAR, etc.), and can also be used quickly for molecular marker assisted selection of powdery mildew resistant gene, and has high accuracy. Combined with other disease resistant gene molecular markers, the identified candidate powdery mildew genes can be used in development of multiresistance breeding material, so as to shorten the breeding period and improve breeding efficiency. The invention lays foundation to elaborate molecular mechanism of powdery mildew resistance of Medicago truncatula.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
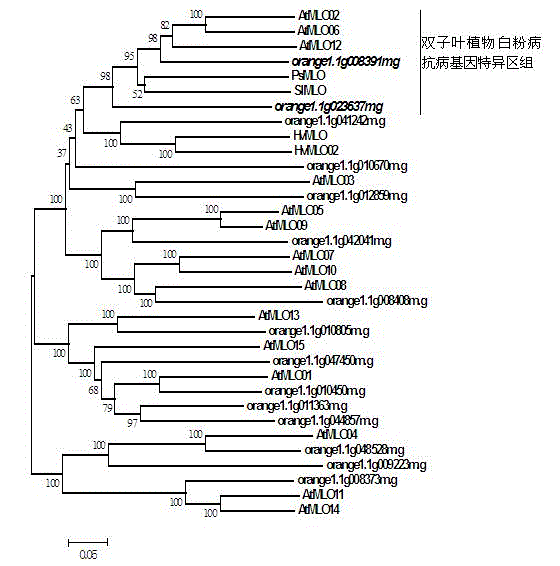
Method for rapidly identifying orange MLO powdery mildew resistance gene
InactiveCN102719446AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesBiotechnologyRapid identification
The present invention is a method for rapidly identifying an orange MLO powdery mildew resistance gene, relating to plant comparative genomics, genetics, bioinformatics and other disciplines of knowledge, and belonging to the field of plant biotechnology science. The method in the invention mainly comprises the following steps of: 1) download of an orange full genome sequence and collection of an MLO gene; 2) identification of the MLO gene; 3) identification of a candidate MLO powdery mildew gene through phylogenetic relationship of the MLO gene; and 4) comparison of the MLO powdery mildew gene. The method in the invention can effectively shorten a mining cycle of the orange powdery mildew gene, facilitating rapid identification of the powdery mildew gene; the method can develop corresponding cosegregation functional markers (SNP, SCAR and the like) through the identified candidate powdery mildew gene and can also be used in fast molecular marker-assisted selection of the powdery mildew resistance gene with high accuracy; the method can also create multiple breeding materials with resistance property through combination of other resistance gene molecular markers, shortening a breeding period and improving breeding efficiency; the method can further lays a foundation for describing an orange powdery mildew resistance molecular mechanism.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
Tracing method of cronobacter spp. in infant formula milk powder
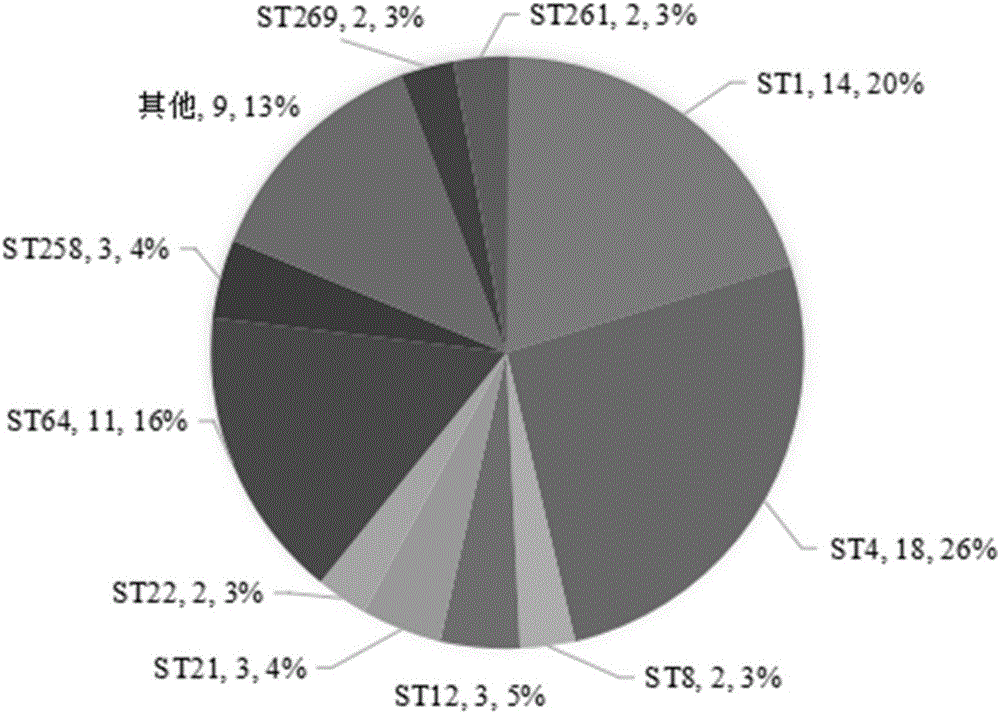
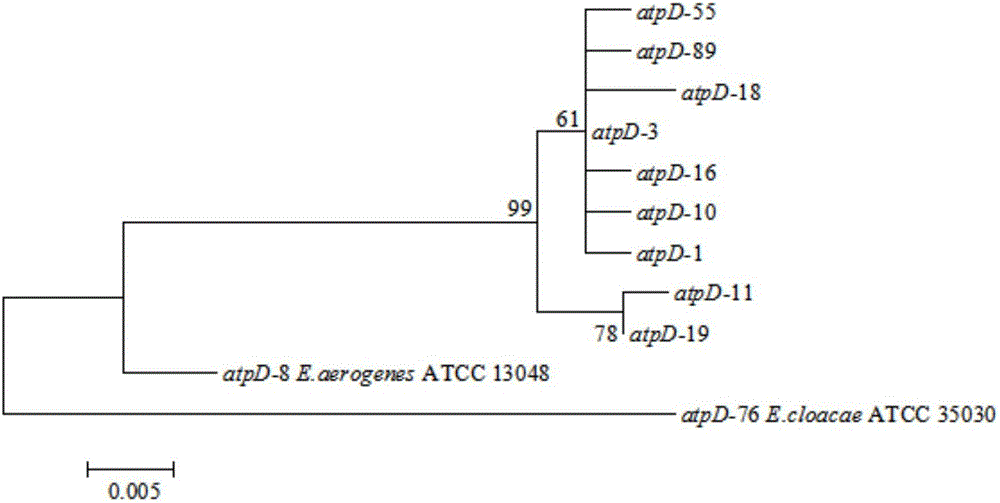
The invention provides a tracing method of cronobacter spp. in infant formula milk powder. The method comprises the steps that by means of a multiple-locus sequence typing technology, molecular typing research is conducted on the cronobacter spp. of PIF raw materials, semi-finished products, finished products and processing environments thereof, population characteristics of the cronobacter spp. are revealed, a phylogenetic relationship of the cronobacter spp. is confirmed, phylogenetic analysis is conducted on the cronobacter spp. through bioinformatics, and therefore genetic characteristics of the cronobacter spp. in the above-mentioned environments are revealed systematically; on the basis of cronobacter spp. MLST typing, correlation analysis is conducted by taking ST of pathogenic bacteria and environmental sources as variables, key links which are most prone to pollute the cronobacter spp. in the PIF processing process and distribution rules are determined, and therefore the source of the cronobacter spp. in the infant formula milk powder is traced. The purpose of fundamentally monitoring the PIF production process and ensuring the PIF quality are achieved.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Method for quickly identifying manihot esculenta mildew-resistance locus (MLO) gene by applying comparative genomics
InactiveCN102796745AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesDiseaseGenetics genomics
The invention discloses a method for quickly identifying a manihot esculenta mildew-resistance locus (MLO) gene, relates to knowledge of plant comparative genomics, genetics, bioinformatics and the like and belongs to the field of plant biotechnology science. The method mainly comprises the following steps of: 1) downloading a manihot esculenta whole genome sequence, and collecting the MLO gene; 2) identifying the MLO gene; 3) identifying the MLO gene according to the MLO gene phylogenetic relationship; and 4) comparing the MLO genes. By the method, the discovery cycle of the manihot esculenta MLO gene is shortened effectively, and the MLO gene can be quickly identified; corresponding coseparation functional markers (single nucleotide polymorphism (SNP) and specific combining ability (SCA)) can be developed by the identified candidate MLO gene, and the method can be quickly used for molecular marker auxiliary selection of the MLO gene, and is high in accuracy; by combining other disease-resistant gene molecular markers, multiresistance breeding materials can be prepared, the breeding cycle is shortened, and the breeding efficiency is improved; and the foundation is laid for elaborating a manihot esculenta MLO gene molecular mechanism.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
Method for establishing phylogenetic tree aiming at target gene of target organism
InactiveCN101962671AMicrobiological testing/measurementSpecial data processing applicationsProtein DatabasesOrganism
The invention relates to a method for establishing a phylogenetic tree aiming at a target gene of a target organism. The method comprises the following steps of: 1) acquiring data; 2) comparing and analyzing sequences; and 3) establishing the phylogenetic tree, wherein the step 1) is finished by downloading a structure domain of a protein of the target gene, acquiring a sequence which contains the structure domain by searching a biological protein database of which genomes and proteomes are sequenced, and searching a protein sequence of a kindred plant by using the target gene of an organism of which the genomes and the proteomes are sequenced. Because the method of the invention comprises the steps and in particular combines the phylogenetic relationships of species with the database of which the genomes and the proteomes are sequenced, a more accurate phylogenetic tree can be established.
Owner:王颖
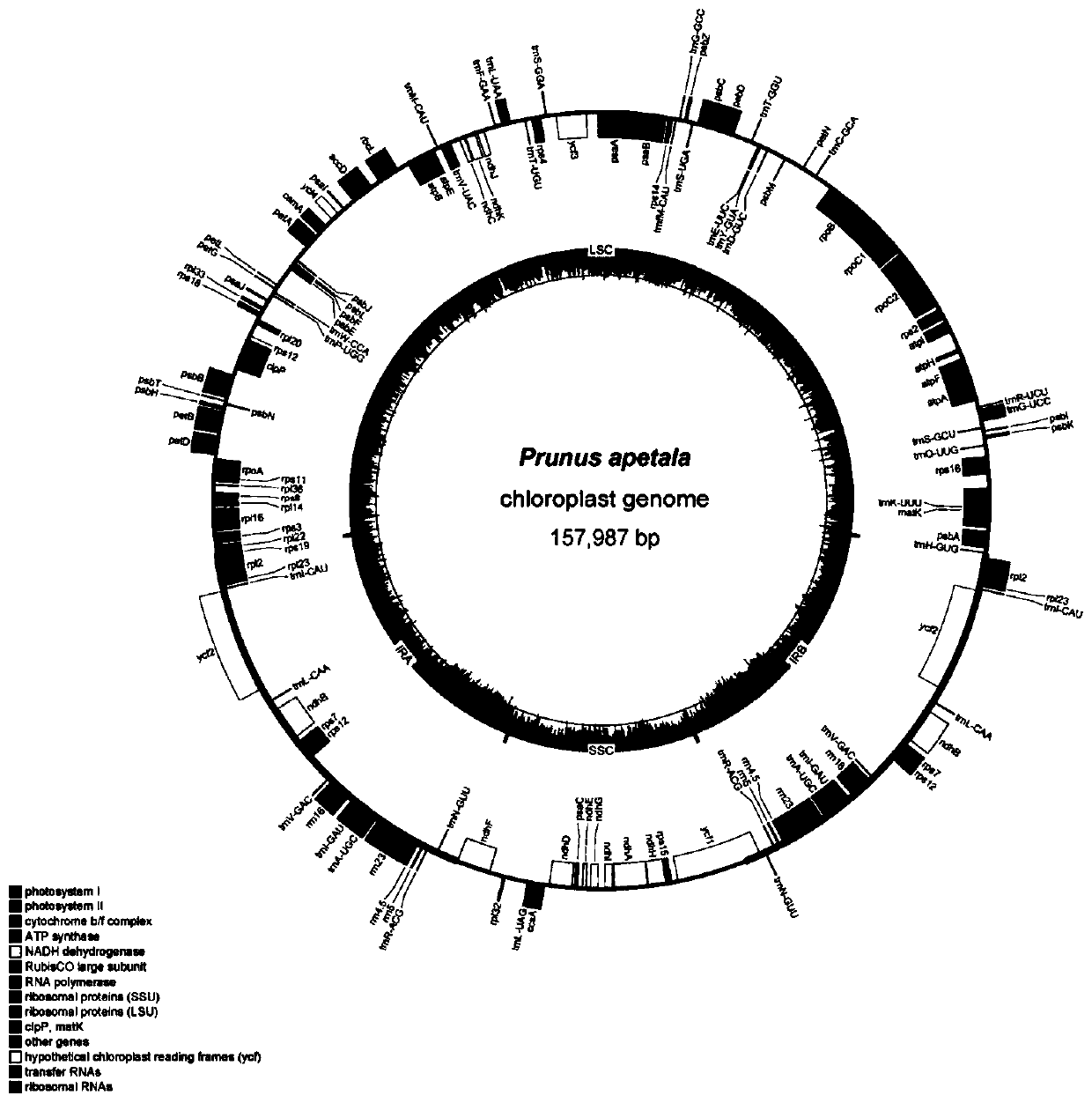
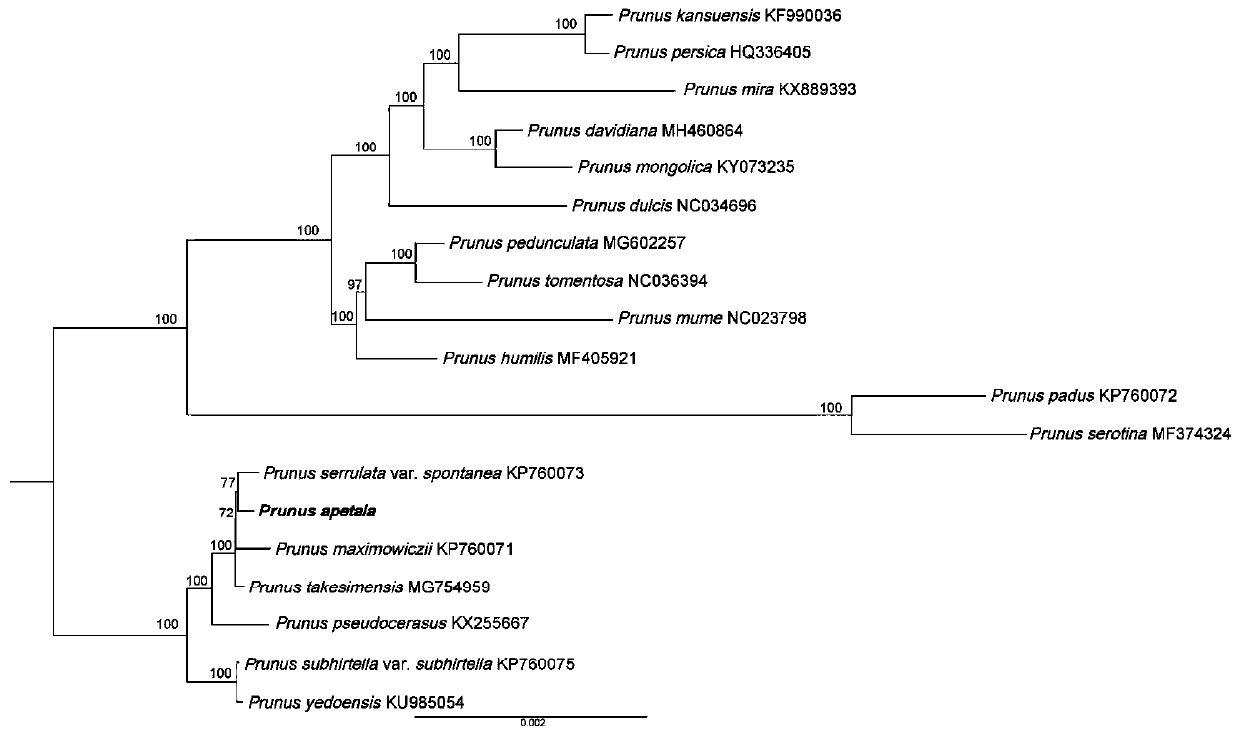
Prunus apetala chloroplast genome and application thereof
The invention discloses a prunus apetala chloroplast genome for the first time. The prunus apetala chloroplast genome is applied to a phylogenetic study of prunus apetala, and a phylogenetic status of the prunus apetala in prunus is analyzed. A phylogenetic relationship of a prunus subgenus species is established through the prunus apetala chloroplast genome by methods of molecular systematics and phylogenetic genomics; on the basis, the whole chloroplast genome is used as a super DNA (deoxyribonucleic acid) bar code of a prunus subgenus for identifying an oriental cherry variety; a technical support is provided for protection of an oriental cherry germplasm resource and species identification; and the chloroplast genome has an important significance for evaluation, protection and utilization of seed breeding and genetic diversity of a typical prunus subgenus plant.
Owner:ZHEJIANG FORESTRY ACAD
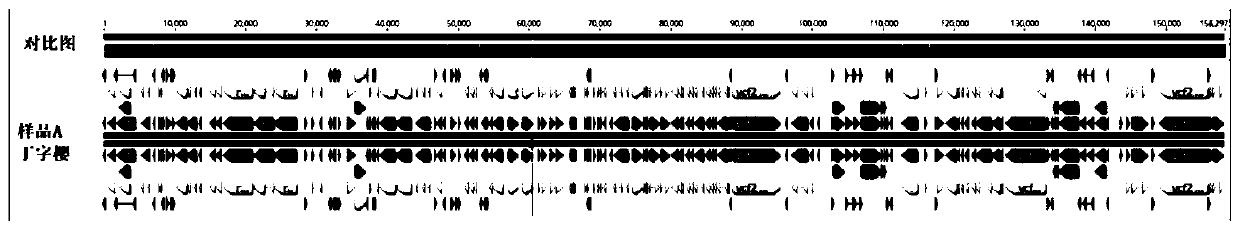
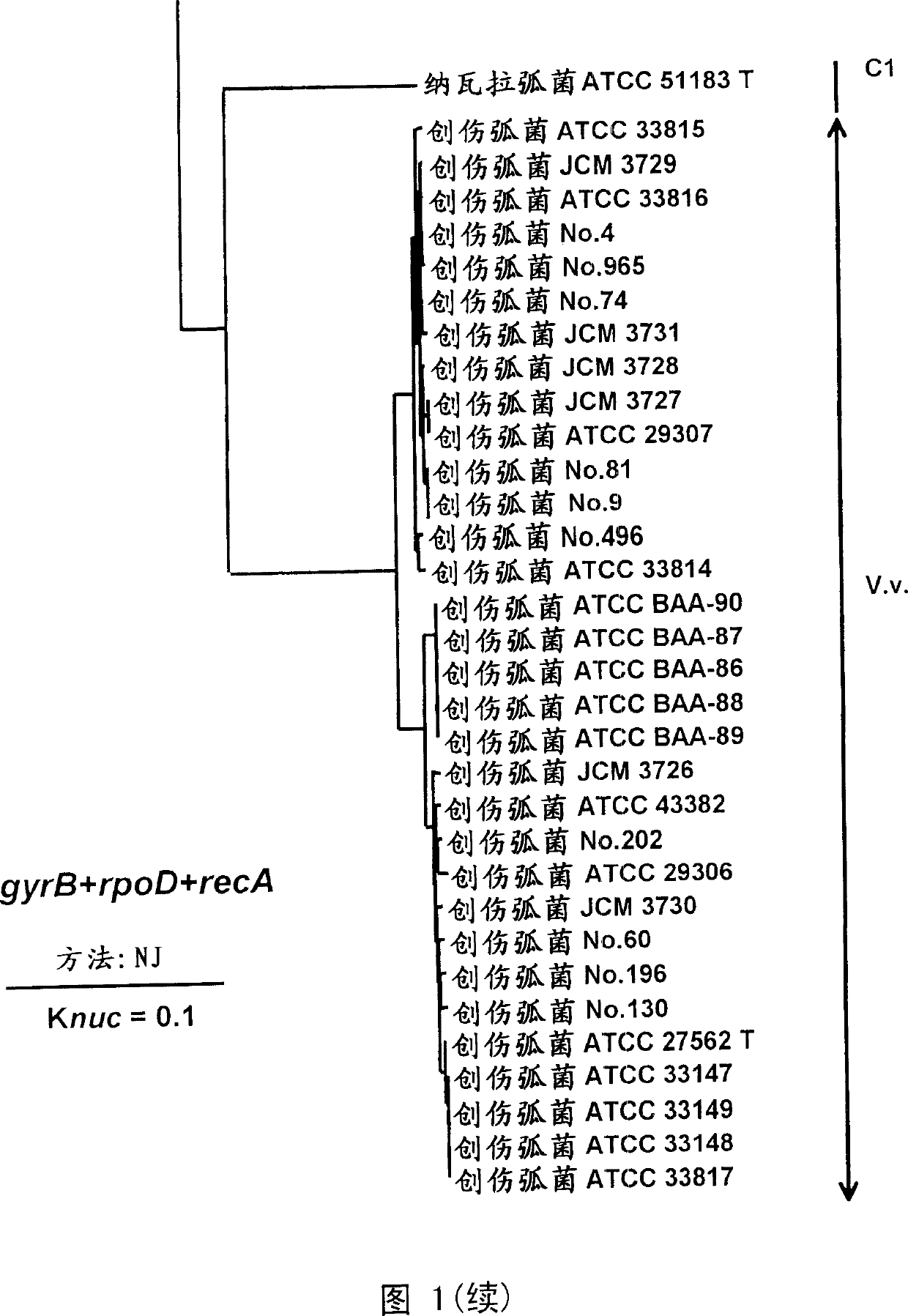
Primer and probe for detecting vibrio vulnificus and detection method using the same
InactiveCN1678754AMicrobiological testing/measurementRecombinant DNA-technologyVibrio vulnificusNucleotide
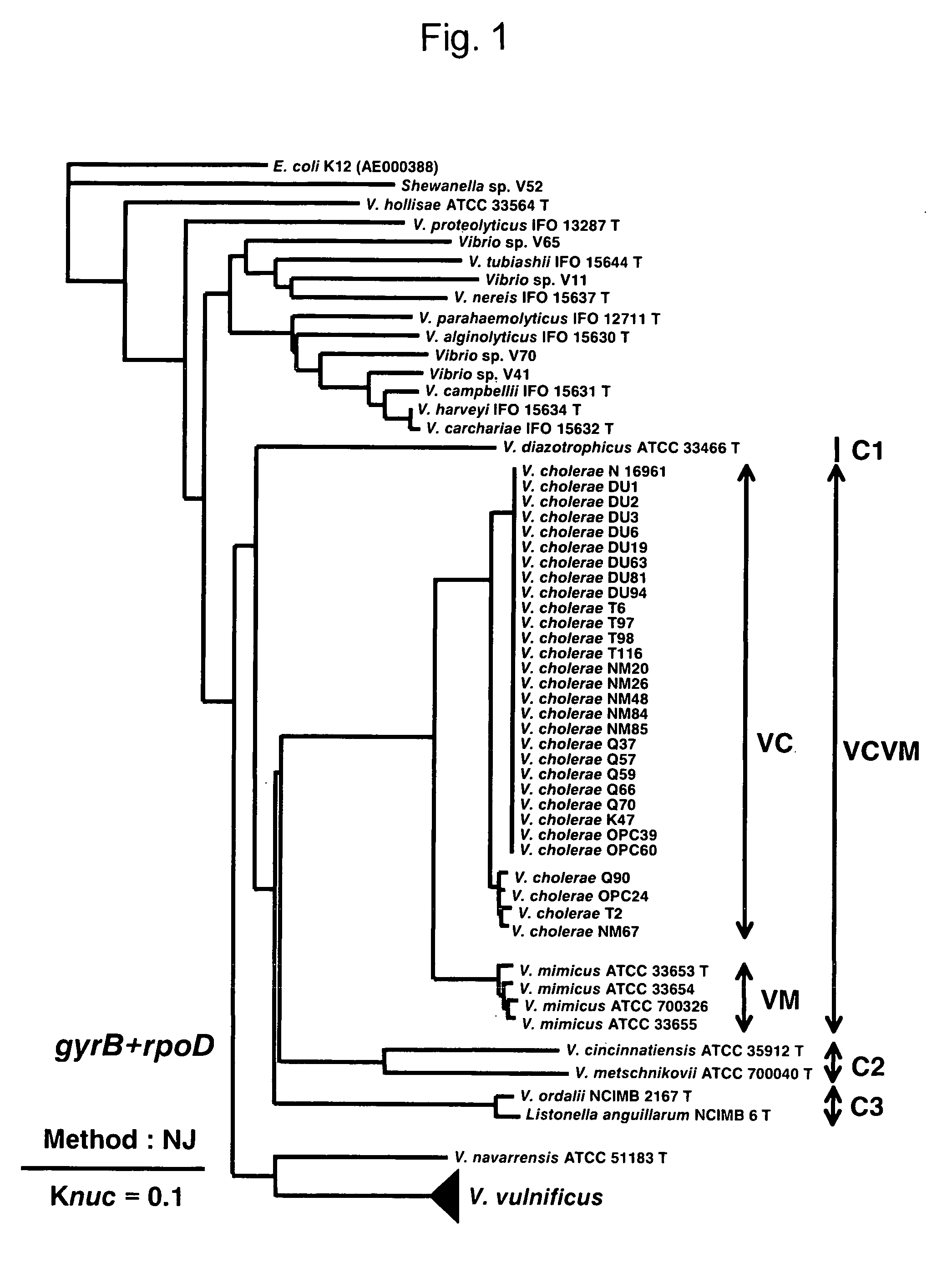
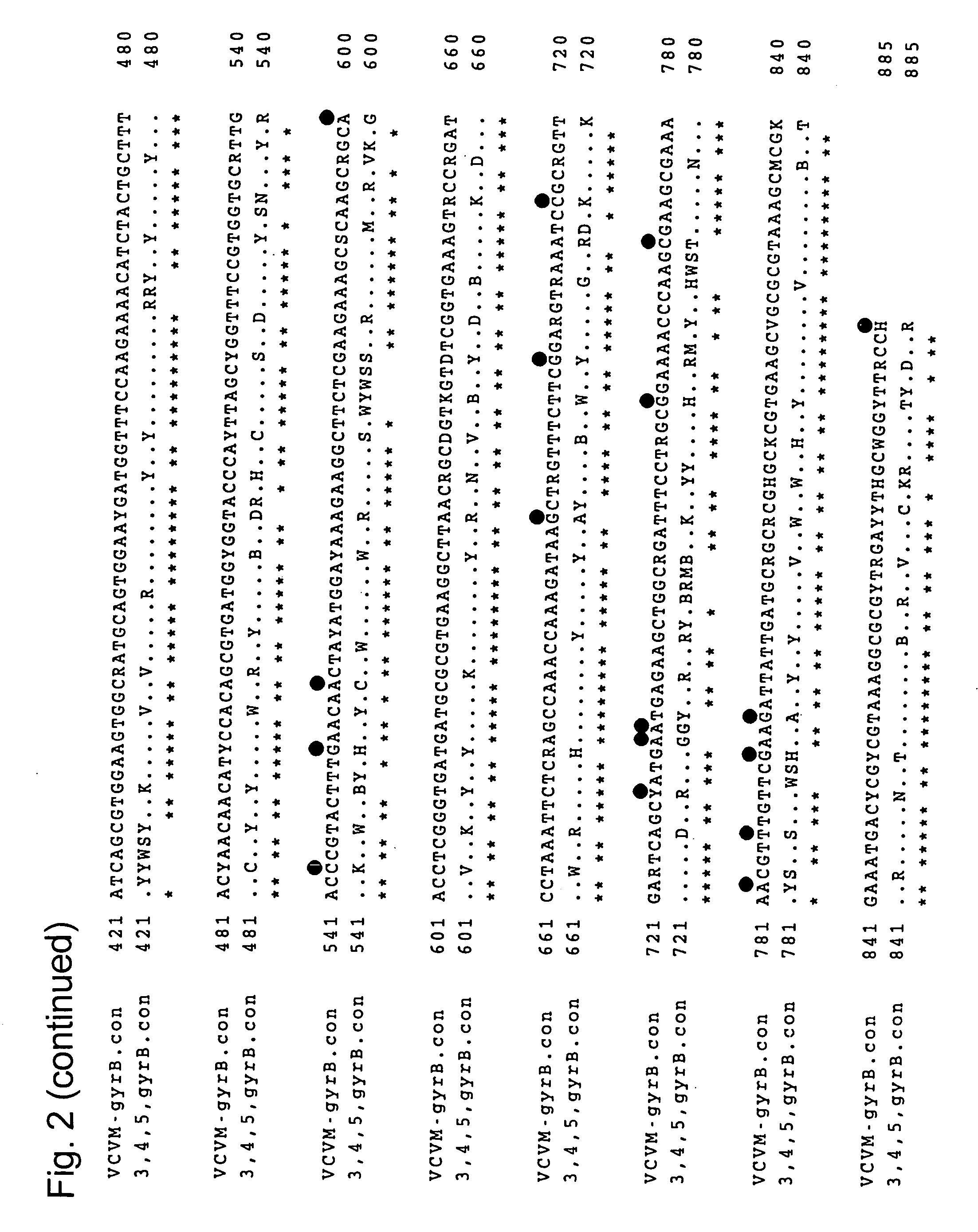
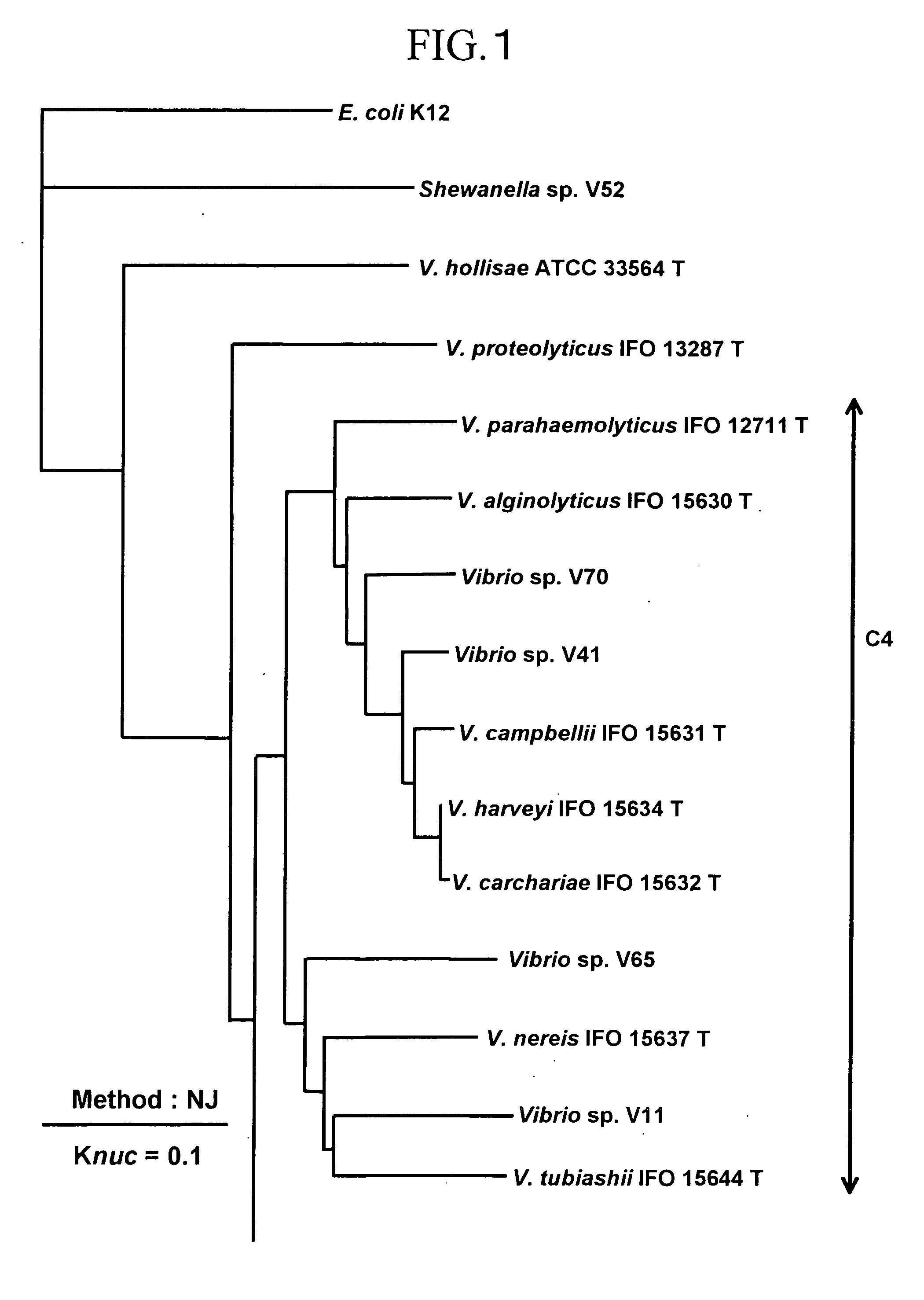
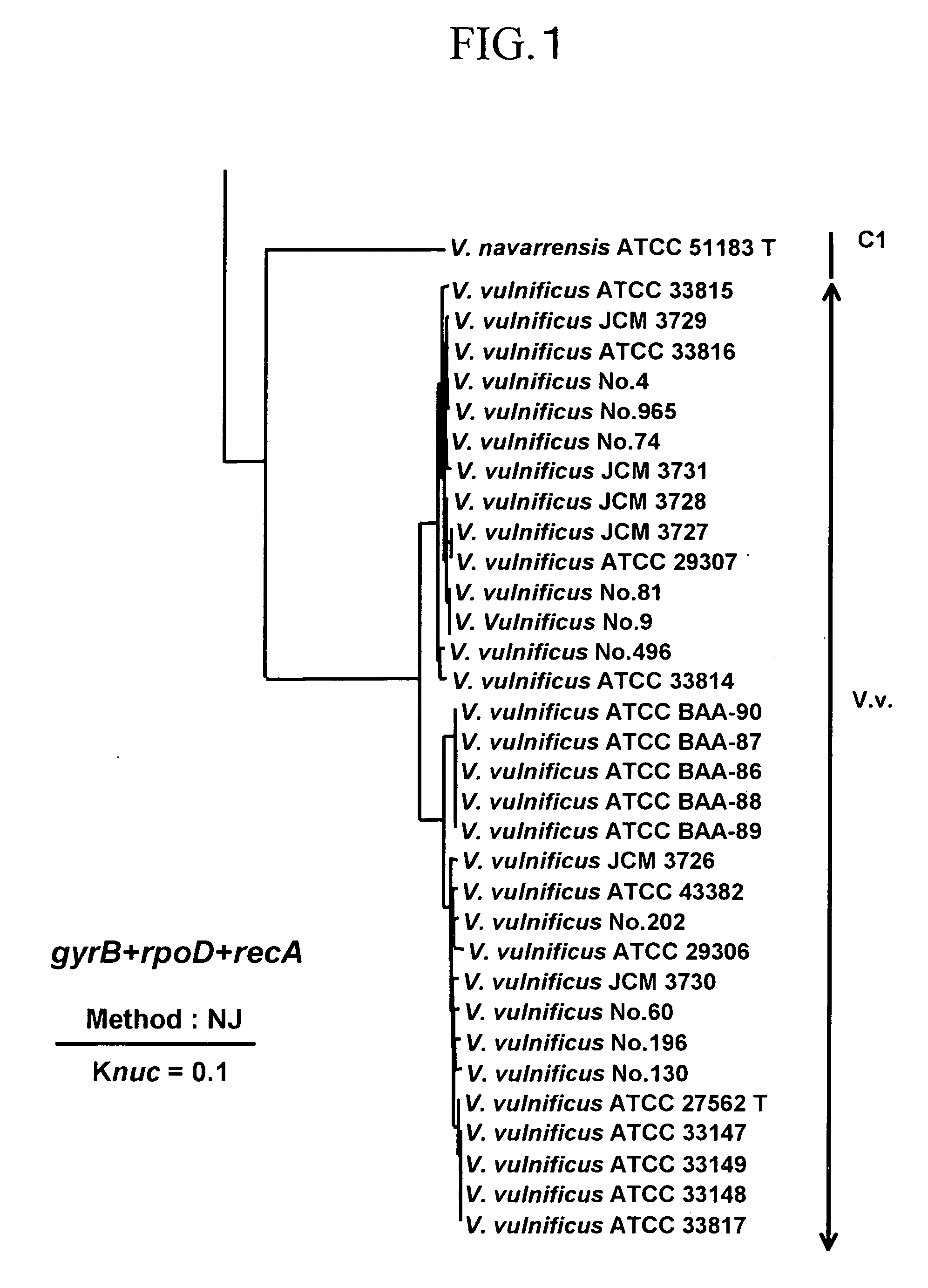
To produce a high-performance specific gene amplification primer for detecting, quantifying, or identifying Vibrio vulnificus, having low risk of misidentification and practically sufficient amplification efficiency and amplification specificity. We have determined partial nucleotide sequences of gyrB, rpoD, and recA genes of Vibrio vulnificus and the closely related species, revealed their phylogenetic relationship, and then identified nucleotides characteristic to Vibrio vulnificus. Thus, we have made it possible to design a probe having high specificity and a gene amplification primer having high specificity and excellent amplification efficiency, both of which contain the characteristic nucleotides.
Owner:NICHIREI CORP
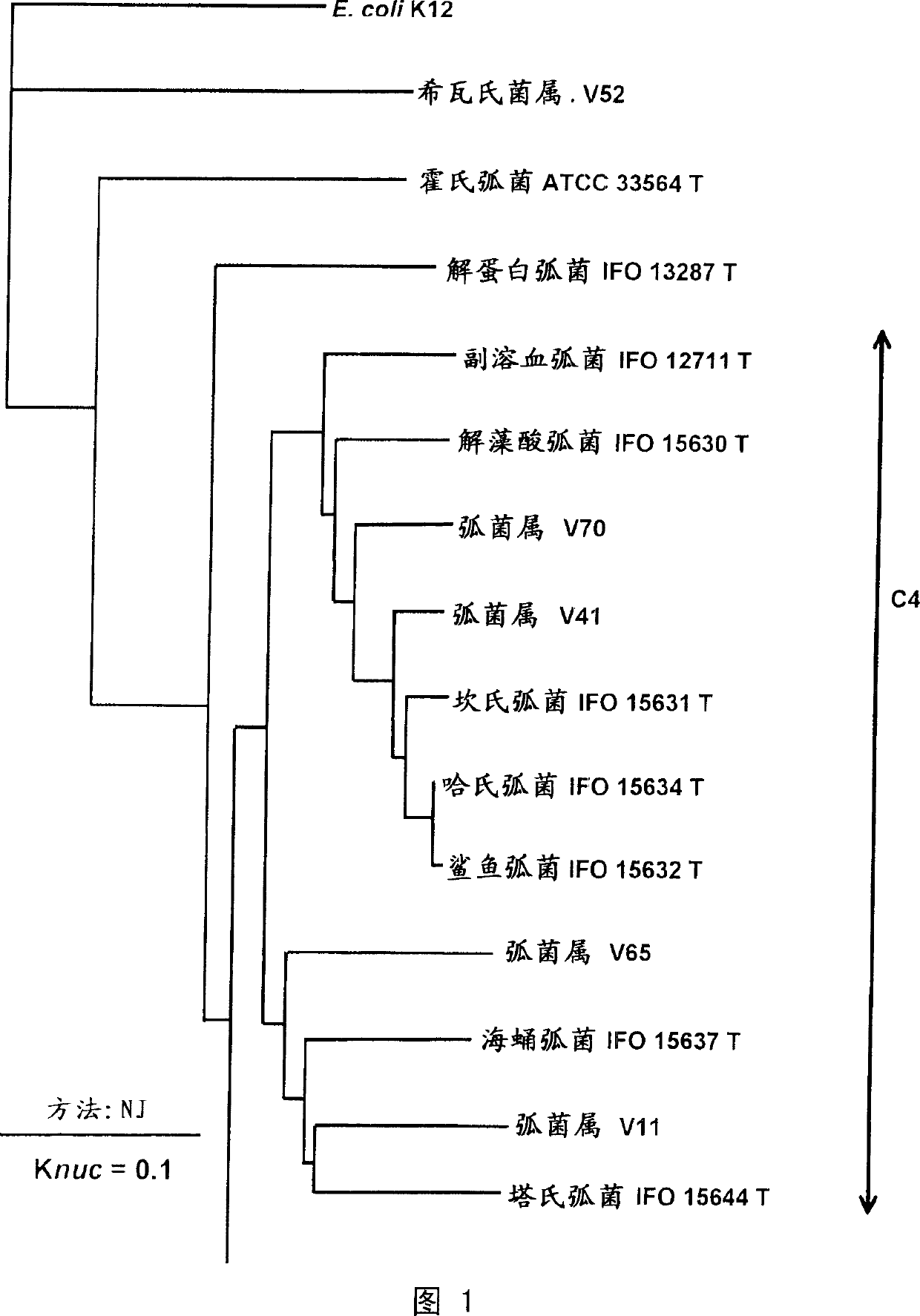
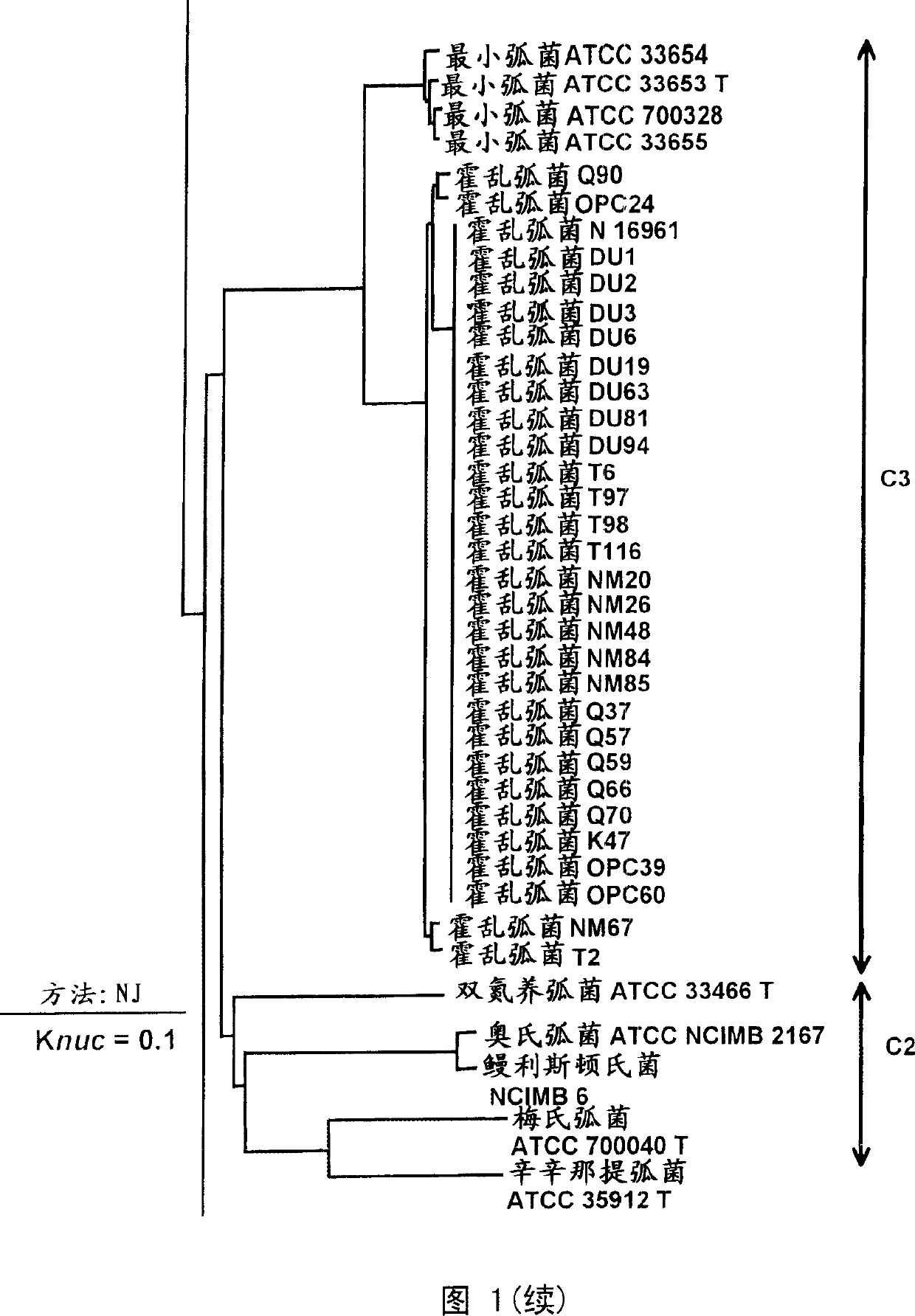
Primer and probe for detecting vibrio cholerae or vibrio mimicus and detection method using the same
InactiveUS20060115820A1Strong specificityHigh amplification efficiencySugar derivativesMicrobiological testing/measurementMicrobiologyNucleotide sequencing
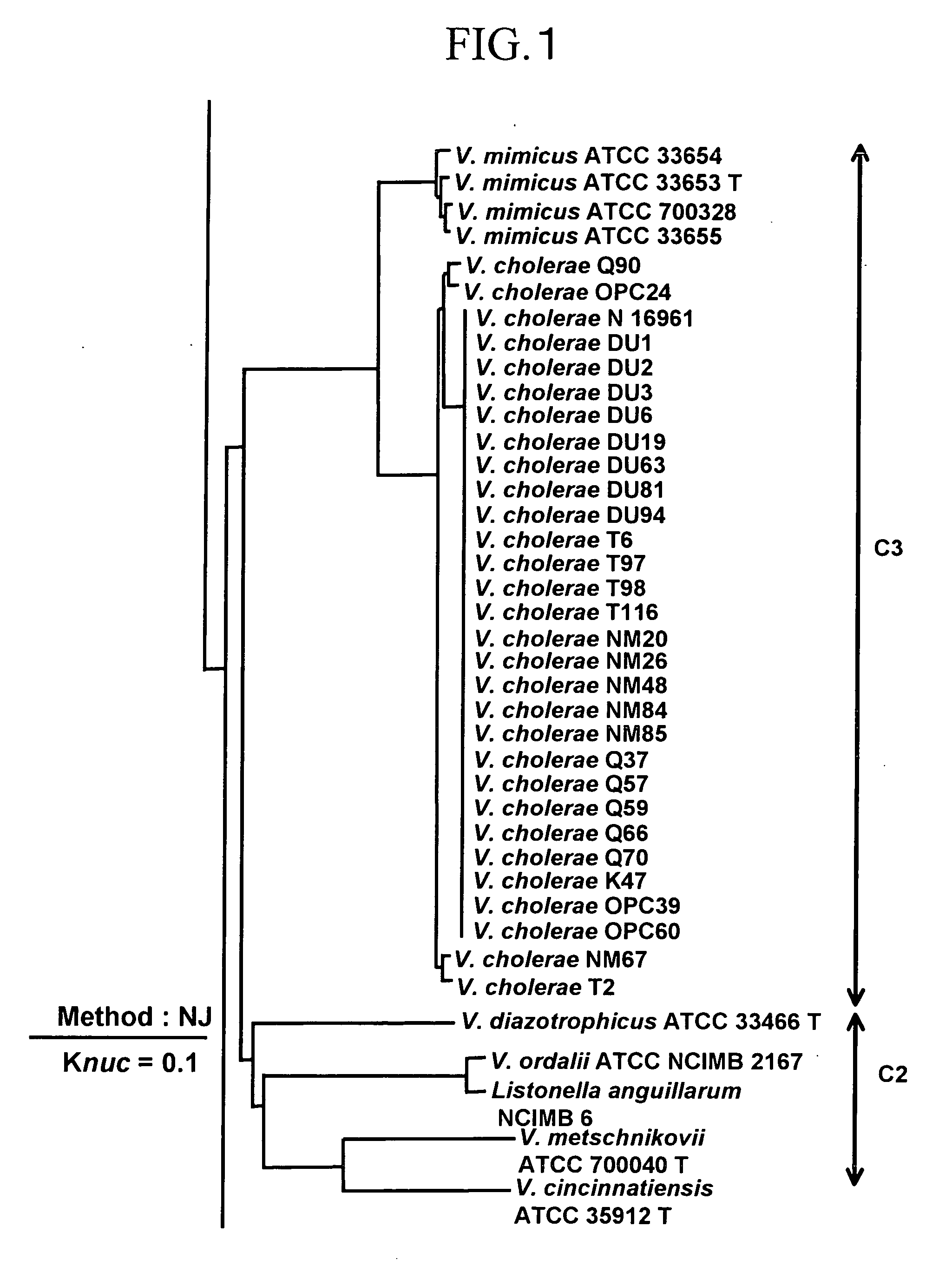
To produce high-performance specific gene amplification primers for detecting, quantifying, or identifying Vibrio cholerae and Vibrio mimicus, having low risk of misidentification and practically sufficient amplification efficiency and amplification specificity. We have determined partial nucleotide sequences of rpoD and gyrB genes of Vibrio cholerae, Vibrio mimicus, and closely related species, revealed their phylogenetic relationship, and then identified nucleotides characteristic of Vibrio cholerae and Vibrio mimicus, respectively. Thus, we have made it possible to design probes having high specificity and gene amplification primers having high specificity and excellent amplification efficiency, both of which contain the characteristic nucleotides.
Owner:NICHIREI FOODSKK
Rapid Identification of pear powdery mildew resistance gene
InactiveCN104561027AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementGenetic engineeringAgricultural scienceRapid identification
The invention relates to rapid identification of a pear powdery mildew resistance gene, and relates to the subject knowledge of plant comparative genomics, genetics and bioinformatics, belonging to the field of science of biotechnology. The rapid identification of the pear powdery mildew resistance gene comprises the following main steps of: 1) downloading of a complete genome sequence of a pear and collection of an MLO gene; 2) identification of the MLO gene; 3) MLO gene phylogenetic relationship; and 4) comparison of an MLO powdery mildew gene. The rapid identification of the pear powdery mildew resistance gene, provided by the invention, has the advantages of effectively shortening the pear powdery mildew gene mining period, and being favorable for the rapid identification of the powdery mildew gene; corresponding co-segregation functional markers (SNP, SCAR and the like) can be developed through the identification of candidate powdery mildew genes, and the co-segregation functional markers can be rapidly used for the molecular marker-assisted selection of the powdery mildew resistance gene, so that the accuracy is high; by combining other powdery mildew resistance gene molecular markers, a multi-resistance breeding material can be created, the breeding period can be shortened, and the breeding efficiency can be improved; a foundation is laid for explaining a pear powdery mildew resistance molecular mechanism.
Owner:CHANGSHU CITY DONGBANG DONGDUN VEGETABLE PROFESSION COOP
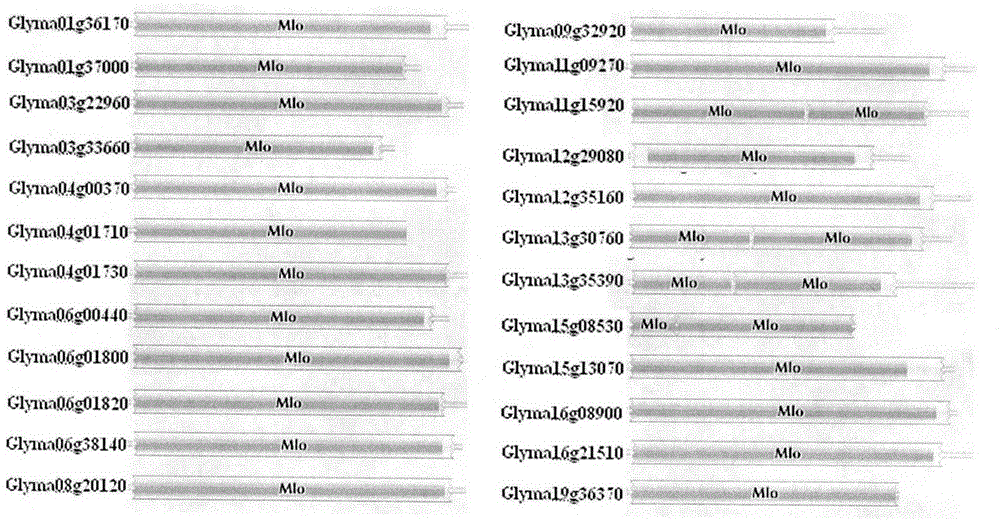
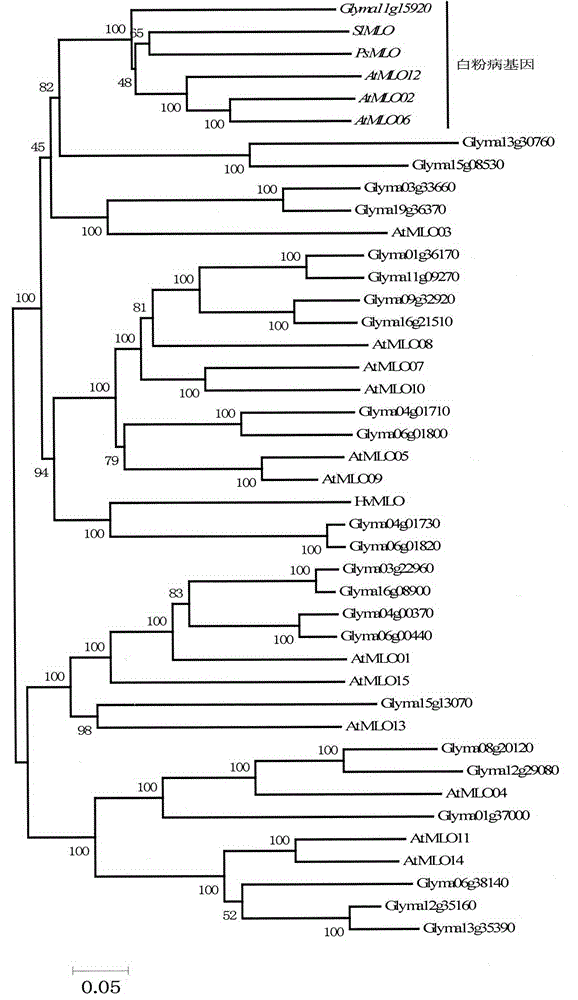
Rapid identification of soybean anti-powdery mildew gene by using candidate gene strategy
InactiveCN104593481AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementBiostatisticsCandidate Gene Association StudyRapid identification
The invention provides rapid identification of soybean anti-powdery mildew gene, relates to knowledge in the disciplines of plant comparative genomics, genetics and bioinformatics, and belongs to the scientific field of plant biotechnology. The invention includes the steps of: 1) downloading the soybean whole genome sequence and acquiring MLO type gene; 2) identifying the MLO genotype; 3) identifying MLO gene phylogenetic relationship; and 4) comparing MLO-type powdery mildew gene. The invention effectively shortens the gene mining cycle for soybean powdery mildew, and is conducive to the rapid identification of anti-powdery mildew gene; corresponding coseparation functional markers (SNP, scar, etc.) are developed through the identified candidate powdery mildew genes, and the identified candidate powdery mildew genes can also be used for fast molecular marker assisted selection of anti-powdery mildew genes and have high accuracy; and combined with other resistant gene molecular markers, multi-resistance breeding material can be created, so as to shorten the breeding period and improve breeding efficiency. Therefore, the invention lays foundation for the explanation of powdery mildew resistant molecular mechanism of soybean.
Owner:JIANGSU CHANGSHU MODERN AGRI IND PARK DEV
Efficient cis-element discovery method using multiple sequence comparisons based on evolutionary relationships
InactiveUS6917883B1Efficient searchBiological testingSpecial data processing applicationsNoncoding DNAPhylogenetic relation
A method is described for efficiently searching non-coding DNA for known control elements. Short conserved DNA sequences can be identified among 3 or more species by selecting DNA sequences from species having a total genetic distance larger than one substitution per site at a neutrally evolving region and calculating the total substitutions using pair-wise genetic distances, with the pairs for comparison selected based on phylogenetic relationships.
Owner:YALE UNIV
Rapid identification of MLO (Mycoplasma Like Organism) powdery mildew resistant genes of melon
InactiveCN104561024AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementFermentationRapid identificationOrganism
The invention relates to rapid identification of MLO (Mycoplasma Like Organism) powdery mildew resistant genes of a melon, relates to comparative genomics of plants, genetics, bioinformatics and other disciplinary knowledge, and belongs to the field of biotechnology science of plants. The rapid identification is mainly carried by the following steps: 1) downloading the whole genome sequence of the melon, and acquiring MLO genes; 2) identifying the MLO genes; 3) obtaining the phylogenetic relationship of MLO genes; 4) comparing MLO powdery mildew resistant genes. By virtue of adopting the rapid identification, the powdery mildew resistant genes mining cycle of the melon can be effectively reduced, which is beneficial to the rapid identification of powdery mildew resistant genes; the identified candidate powdery mildew resistant genes are utilized to develop corresponding coseparation functional markers (such as SNR and SCAR) and can also be quickly used for assistant selection of molecular markers resistant to powdery mildew resistant genes, and the accuracy is high; in combination with other anti-disease gene molecular markers, a plurality of resistive breeding materials can be created, the breeding periods can be reduced, and the breeding efficiency can be increased; the foundation is provided for describing a powdery mildew resistant molecular mechanisms of the melon.
Owner:南农大(常熟)新农村发展研究院有限公司
Amphibian mitochondria general macro bar code amplification primers and application method thereof
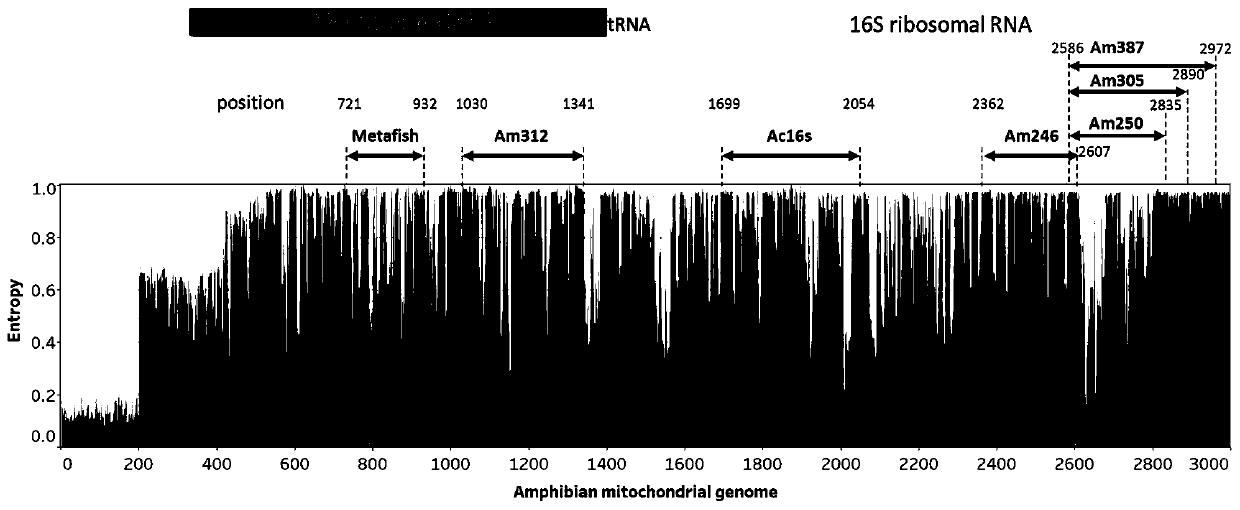
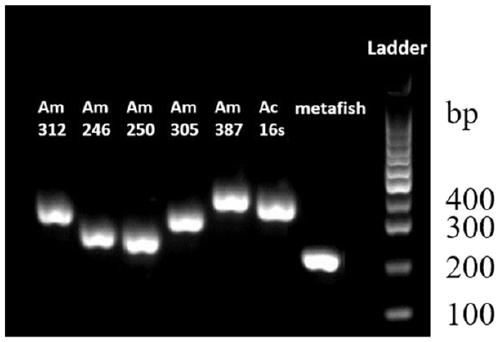
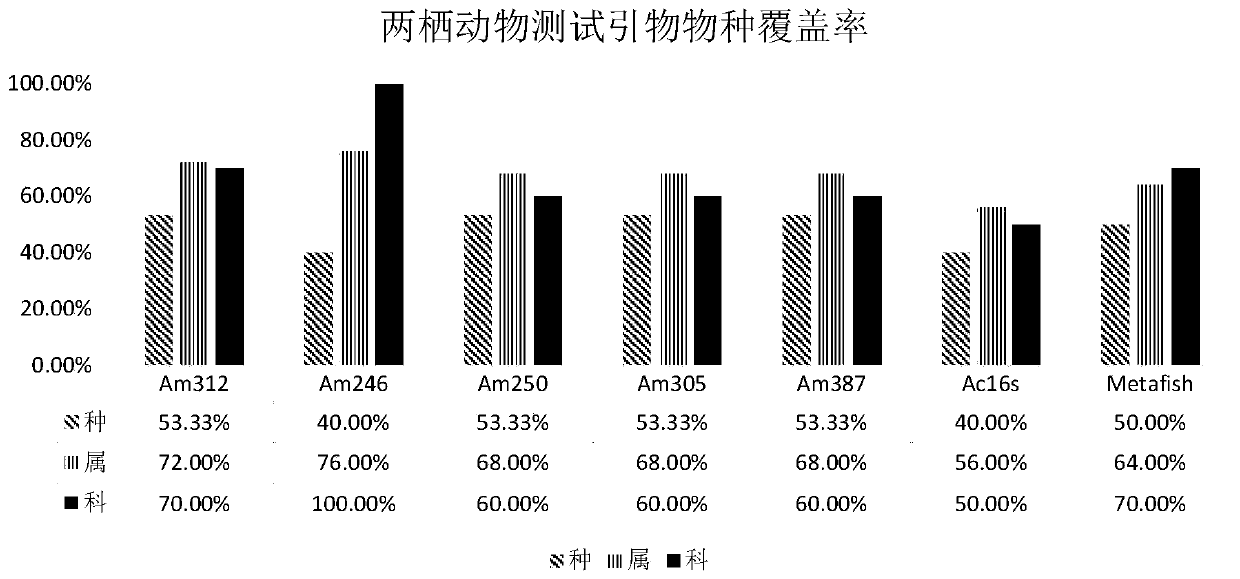
PendingCN111549146AImprove general performanceGuaranteed resolutionMicrobiological testing/measurementDNA/RNA fragmentationForward primerPhylogenetic relationship
The invention belongs to the technical field of gene detection, and discloses amphibian mitochondria general macro bar code amplification primers and an application method thereof. The amplification primers comprise a primer pair formed by combining at least one forward primer and one reverse primer. Products obtained by PCR amplification through combination of Am312-F1 and Am312-R1, combination of Am246-F2 and Am246-R2, combination of Am250-F3 and Am250-R3.1, combination of Am250-F3 and Am305-R3.2 and combination of Am250-F3 and Am387-R3.3 meet the requirements of next-generation sequencing and can be used for species identification and diversity analysis of amphibians; through flexible combination of the above primer pairs, the research purposes of amphibian species classification, phylogenetic relationship, systematic geography and the like can be achieved.
Owner:南京易基诺环保科技有限公司
Primer and probe for detecting vibrio vulnificus and detection method using the same
InactiveUS20070054267A1Reduce the possibilitySufficient amplification efficiencySugar derivativesMicrobiological testing/measurementVibrio vulnificusNucleotide sequencing
To produce a high-performance specific gene amplification primer for detecting, quantifying, or identifying Vibrio vulnificus, having low risk of misidentification and practically sufficient amplification efficiency and amplification specificity. We have determined partial nucleotide sequences of gyrB, rpoD, and recA genes of Vibrio vulnificus and the closely related species, revealed their phylogenetic relationship, and then identified nucleotides characteristic to Vibrio vulnificus. Thus, we have made it possible to design a probe having high specificity and a gene amplification primer having high specificity and excellent amplification efficiency, both of which contain the characteristic nucleotides.
Owner:NICHIREI CORP
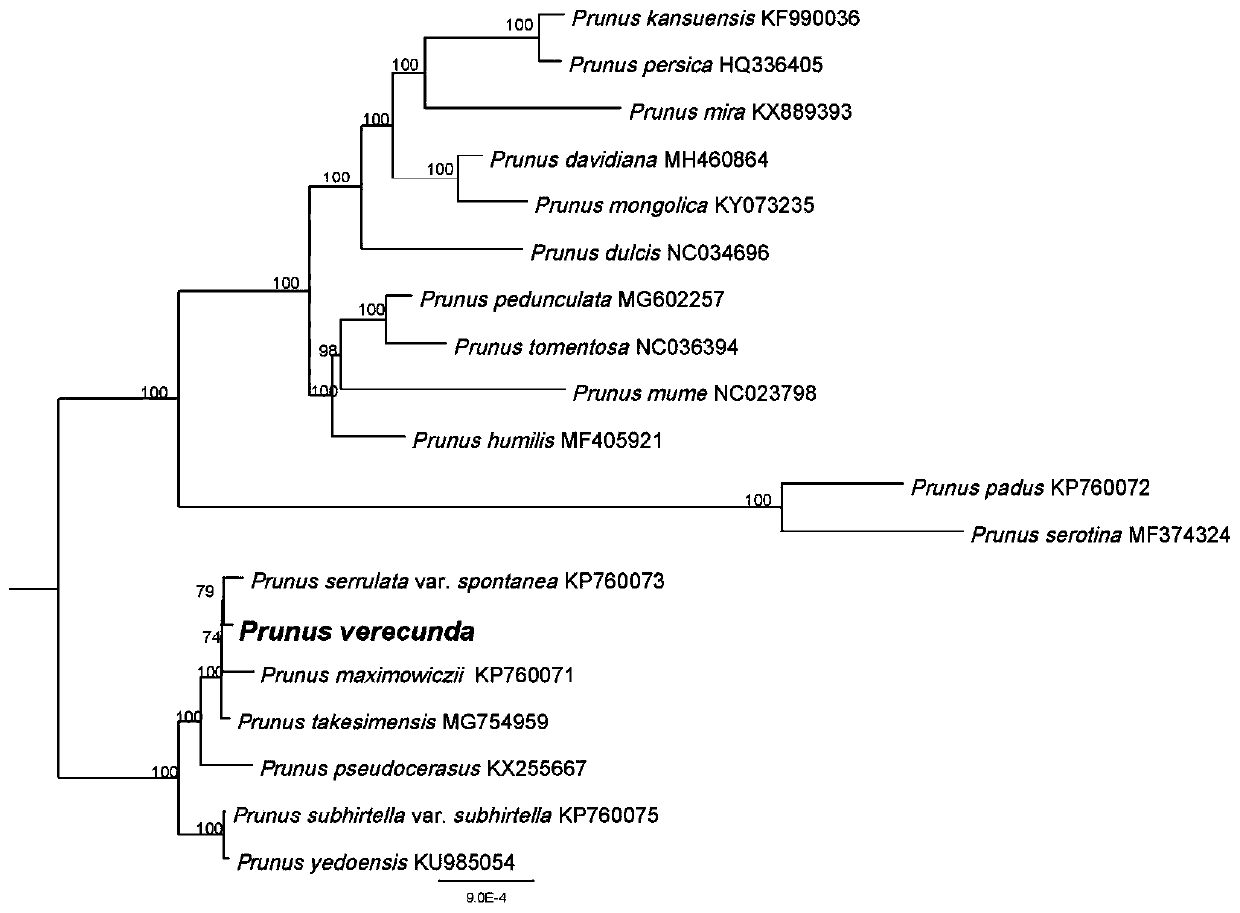
Prunus verecunda chloroplast genome and application thereof
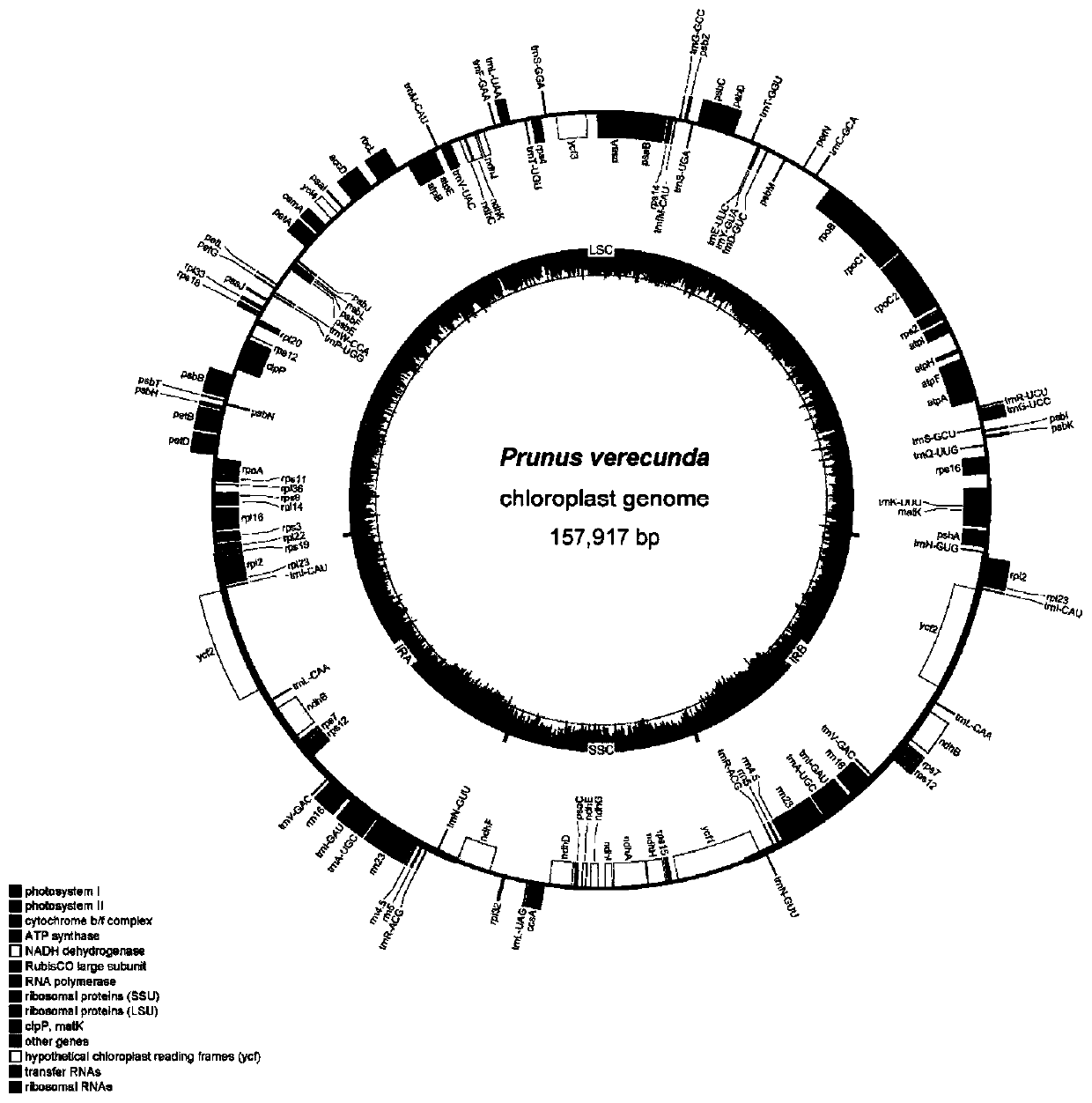
ActiveCN110305877AParsing phylogenetic positionMicrobiological testing/measurementPlant peptidesGenomicsPhylogenetic study
The invention discloses a prunus verecunda chloroplast genome for the first time, applies the prunus verecunda chloroplast genome to prunus verecunda system growth research, and analyzes the prunus verecunda growth status in prunus. According to the prunus verecunda chloroplast genome, molecular systematics and phylogeny genomics method are used for establishing phylogenetic relationship of subgenus cerasus species through the chloroplast genome, on the basis, the whole chloroplast genome is taken as a super DNA bar code of subgenus cerasus and used for identifying sakura varieties, technicalsupports for protecting of sakura germplasm resources and species identification are provided, and great significances on breeding typical subgenus cerasus plants, and the evaluation, protection and utilization of genetic diversity are brought.
Owner:ZHEJIANG FORESTRY ACAD
Application of comparative genomics to rapid identification of phaseolus vulgaris mildew resistance locus o gene
InactiveCN104593480AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementBiostatisticsRapid identificationOrganism
Relating to plant comparative genomics, genetics, bioinformatics and other disciplinary knowledge, the invention belongs to the scientific field of plant biotechnology, and discloses rapid identification of phaseolus vulgaris mildew resistance locus o gene. The main steps include: 1) download of phaseolus vulgaris whole genome sequence and acquisition of MLO type gene; 2) identification of MLO type gene; 3) MLO type gene phylogenetic relationship; and 4) comparison of MLO type powdery mildew. The invention effectively shortens the mining cycle of phaseolus vulgaris powdery mildew gene, and is conducive to rapid identification of powdery mildew gene. Corresponding coseparation functional markers (SNP, SCAR and the like) are developed through identified candidate powdery mildew genes, which also can be used for molecular marker-assisted selection of mildew resistance locus o gene and have high accuracy. Multi-resistance breeding materials can be created by combining other resistant gene molecular markers, the breeding period can be shortened, and the breeding efficiency is improved, thus laying the foundation for elaborating the phaseolus vulgaris mildew resistance molecular mechanism.
Owner:JIANGSU CHANGSHU MODERN AGRI IND PARK DEV
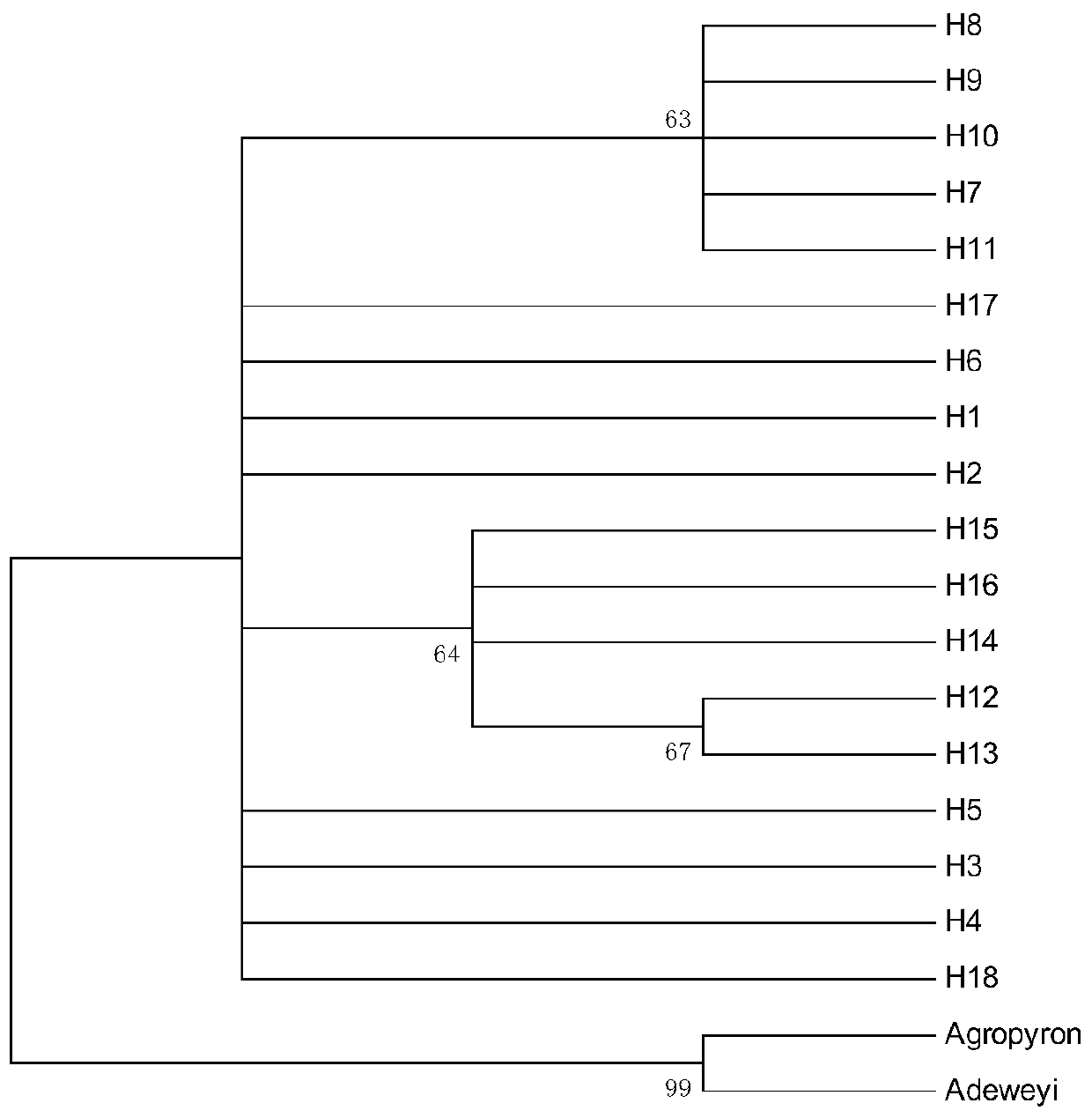
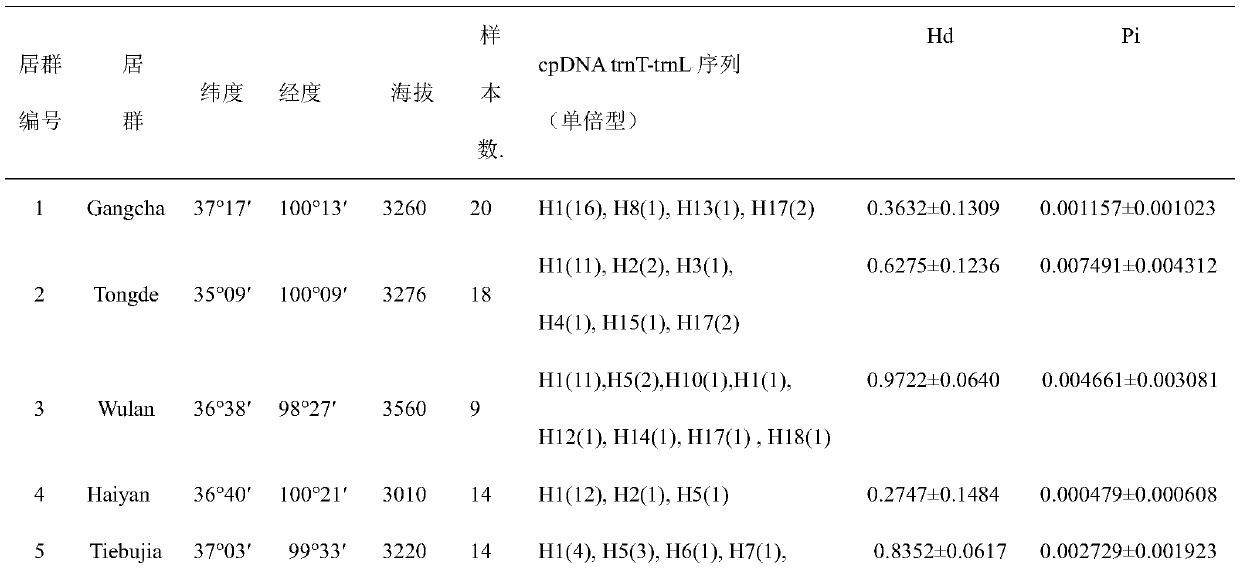
Method for researching polymorphism of different geographical populations cpDNatnT-trnL of Snowfox Crested wheatgrass
InactiveCN110060732AAccurately capture levels of genetic variationAccurately obtain population genetic informationMicrobiological testing/measurementBiostatisticsNucleotide sequencingSequence variation
The invention discloses a method for researching polymorphism of different geographical populations cpDNatnT-trnL of Snowfox Crested wheatgrass. The method includes the steps: collecting a plurality of tender leaves of the Snowfox Crested wheatgrass in a wheatgrass population; extracting the total DNA of each tender leaf of the Snowfox Crested wheatgrass; amplifying the trnT-trnL fragment of the cpDNA of each tender leaf of the Snowfox Crested wheatgrass to obtain a plurality of amplification products; carrying out sequencing to obtain a nucleotide sequence of a trnT-trnL amplified fragment ofthe cpDNA of each tender leaf of the Snowfox Crested wheatgrass; analyzing the sequence data by adopting software; and according to data analysis, obtaining cpDNAtrnT-trnL polymorphism information ofthe Snowfox Crested wheatgrass in different geographical populations, including sequence variation, genetic diversity, genetic structure of populations and phylogenetic relationship information amongthe populations. The method for researching polymorphism of different geographical populations cpDNatnT-trnL of Snowfox Crested wheatgrass can accurately obtain the polymorphism information of different geographical populations of the Snowfox Crested wheatgrass, and can provide a scientific basis for species protection.
Owner:青海省铁卜加草原改良试验站
Rapid identification of powdery mildew resistant gene of Chinese cabbage by using comparative genomics
InactiveCN104593379AShorten digging cycleRapid identificationMicrobiological testing/measurementFermentationRapid identificationGenotype
The invention provides rapid identification of powdery mildew resistant gene of Chinese cabbage, relates to knowledge in the disciplines of plant comparative genomics, genetics and bioinformatics, and belongs to the scientific field of plant biotechnology. The invention includes the steps of: 1) downloading the whole genome sequence of Chinese cabbage and acquiring MLO type gene; 2) identifying the MLO genotype; 3) identifying MLO gene phylogenetic relationship; and 4) comparing MLO-type powdery mildew gene. The invention effectively shortens the gene mining cycle for Chinese cabbage powdery mildew, and is conducive to the rapid identification of powdery mildew resistant gene; corresponding coseparation functional markers (SNP, SCAR, etc.) are developed through the identified candidate powdery mildew genes, and the identified candidate powdery mildew genes can also be used for fast molecular marker assisted selection of powdery mildew resistant genes and have high accuracy; combined with other resistant gene molecular markers, multi-resistance breeding material can be created, so as to shorten the breeding period and improve breeding efficiency. Therefore, the invention lays foundation for the explanation of powdery mildew resistant molecular mechanism of Chinese cabbage.
Owner:JIANGSU CHANGSHU MODERN AGRI IND PARK DEV
Specific SSR primers for malus sieversii and cultivated apple and applications of specific SSR primers
ActiveCN108660239AHigh differentiation levelLow level of differentiationMicrobiological testing/measurementBiostatisticsAgricultural scienceMalus sieversii
The invention discloses specific SSR primers for malus sieversii and the cultivated apple and applications of the specific SSR primers. Based on the whole genome sequence of the cultivated apple, a functional gene sequence relevant to the stress resistance of apple is adopted as a template, and 15 pairs of SSR primers amplifying SSR target bands with specificity, stability and polymorphism in malus sieversii and the cultivated apple are screened, wherein the nucleotide sequences of the SSR primers are as shown in SEQ ID No.1-30. 8 pairs of SSR primers only capable of amplifying SSR target bands in the cultivated apple with specificity are also screened. The protein products coded by the target bands amplified by the SSR primers are known, and the SSR primers can be used for carrying out genetic diversity, polymorphis or phylogenetic analysis on the malus sieversii and cultivated apple, thus disclosing the genetic diversity and phylogenetic relationship of the malus sieversii and cultivated apple and the influences of the obtained genetic variation on the malus sieversii and cultivated apple in the aspects including the biological function.
Owner:INST OF FOREST ECOLOGY ENVIRONMENT & PROTECTION CHINESE ACAD OF FORESTRY
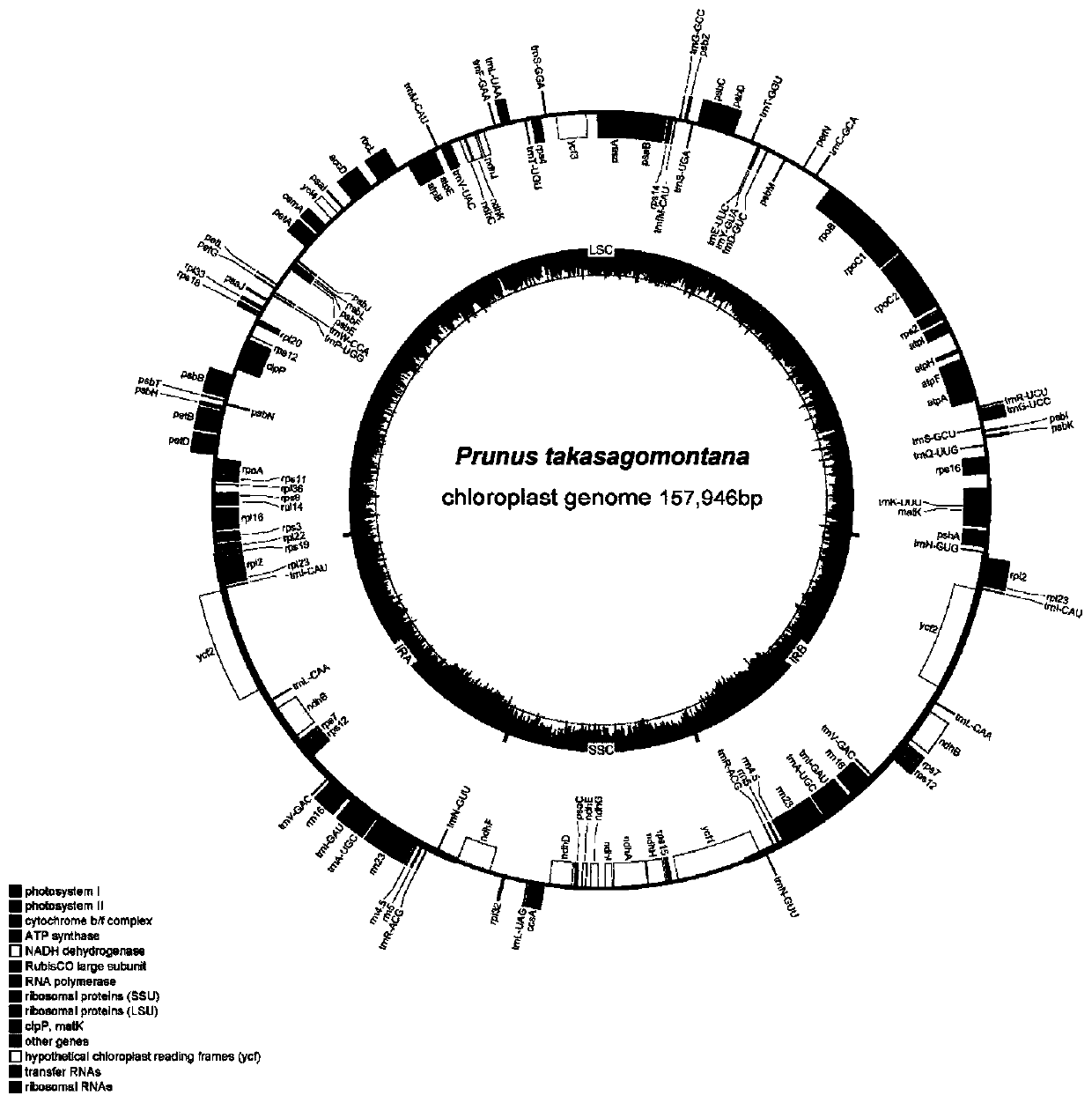
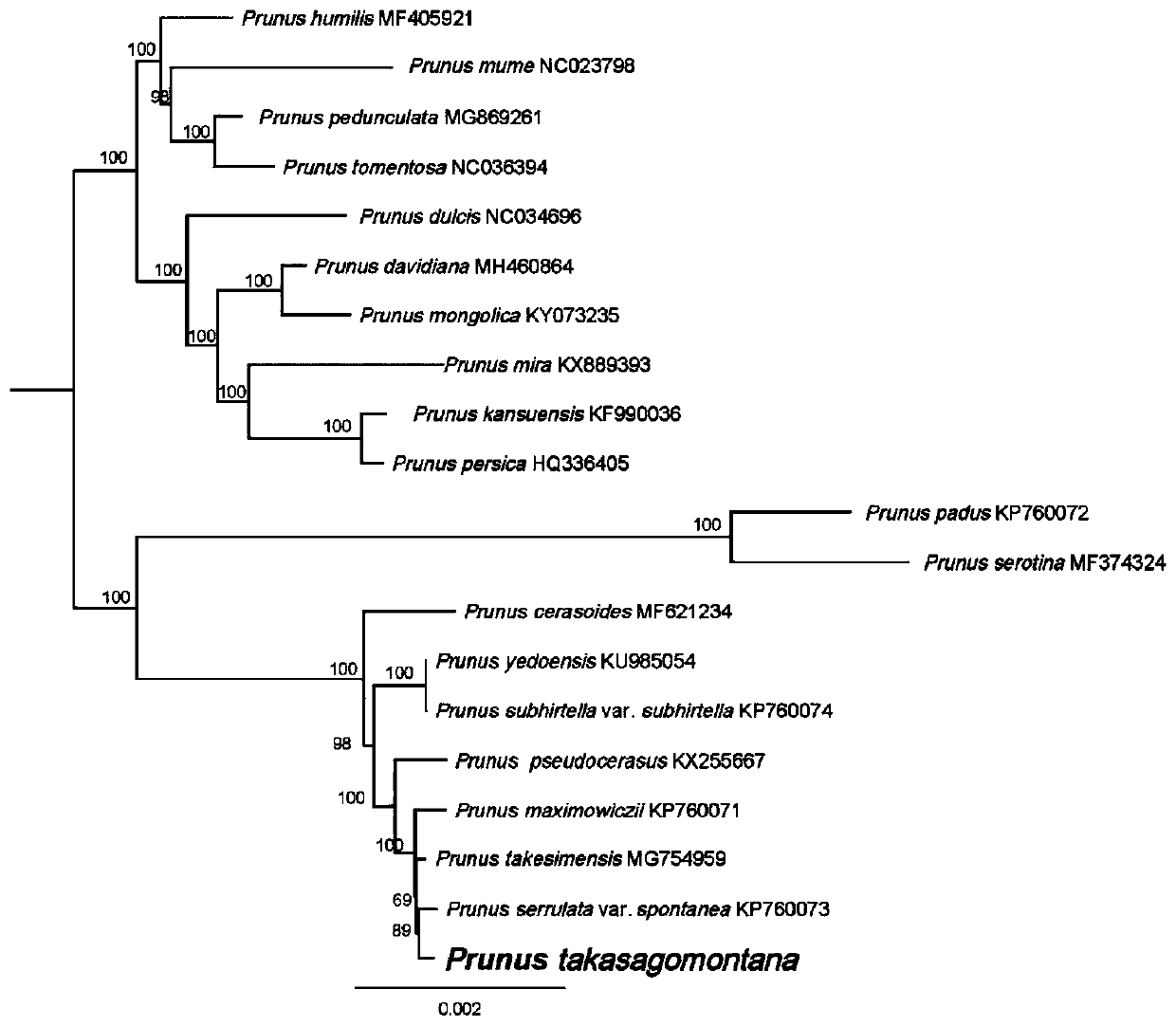
Prunus takasagomontan chloroplast genome and application thereof
ActiveCN110358857AParsing phylogenetic positionMicrobiological testing/measurementData visualisationPhylogenetic studyGenomics
The invention discloses a prunus takasagomontan chloroplast genome for the first time. The prunus takasagomontan chloroplast genome is applied to the phylogenetic study of prunus takasagomontan to analyze the phylogenetic status of the prunus takasagomontan in prunus. According to the study, molecular systematics and phylogenetic genomics methods are utilized, a phylogenetic relationship of subgenus cerasus species is established through the prunus takasagomontan chloroplast genome, and on this basis, the whole chloroplast genome is utilized as a super DNA bar code of the subgenus cerasus foridentifying varieties of cerasus, which provides technical support for protection and species identification of cerasus germplasm resources and is of great significance for breeding, genetic diversityevaluation, protection and utilization of typical subgenus cerasus plants.
Owner:ZHEJIANG FORESTRY ACAD
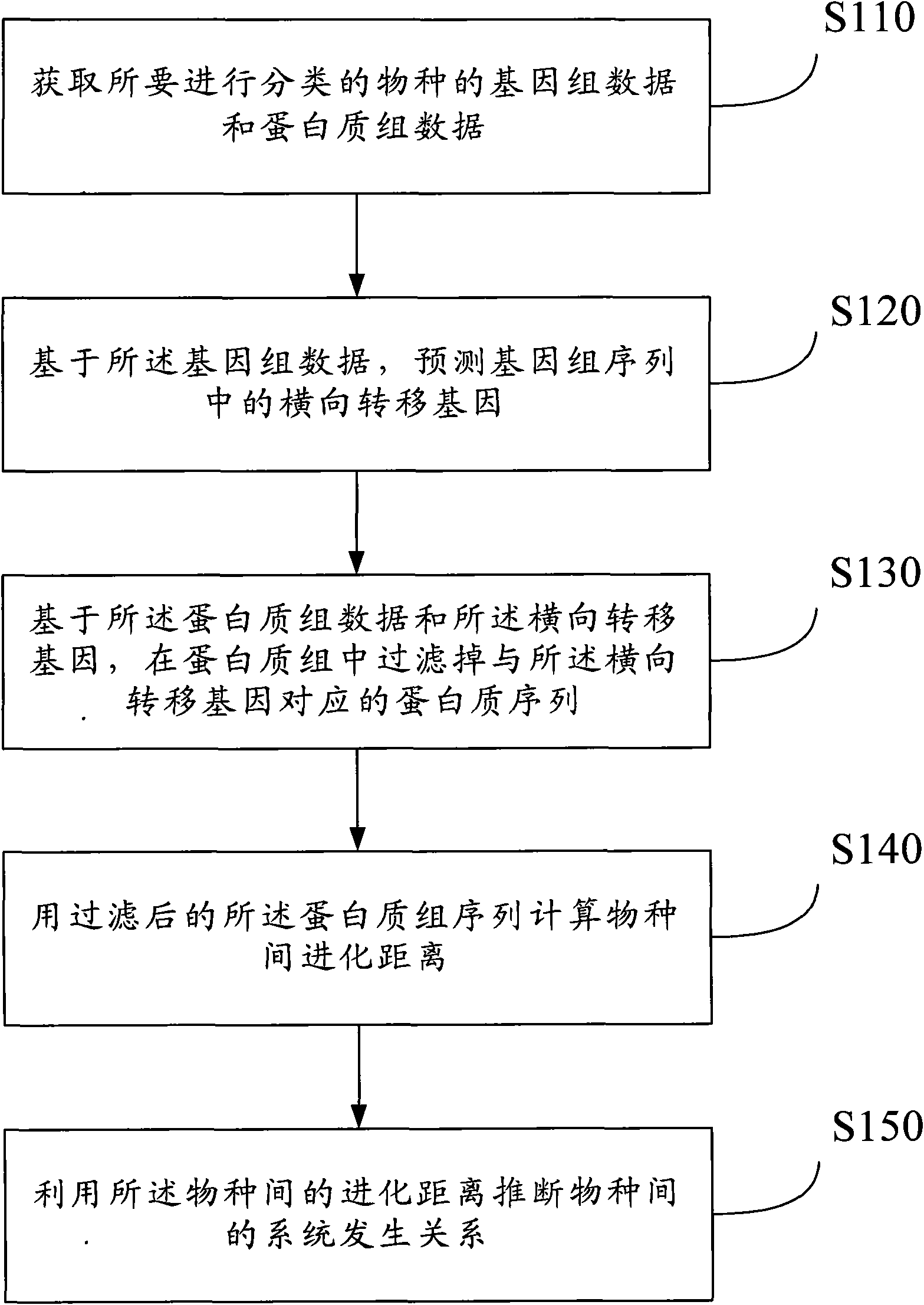
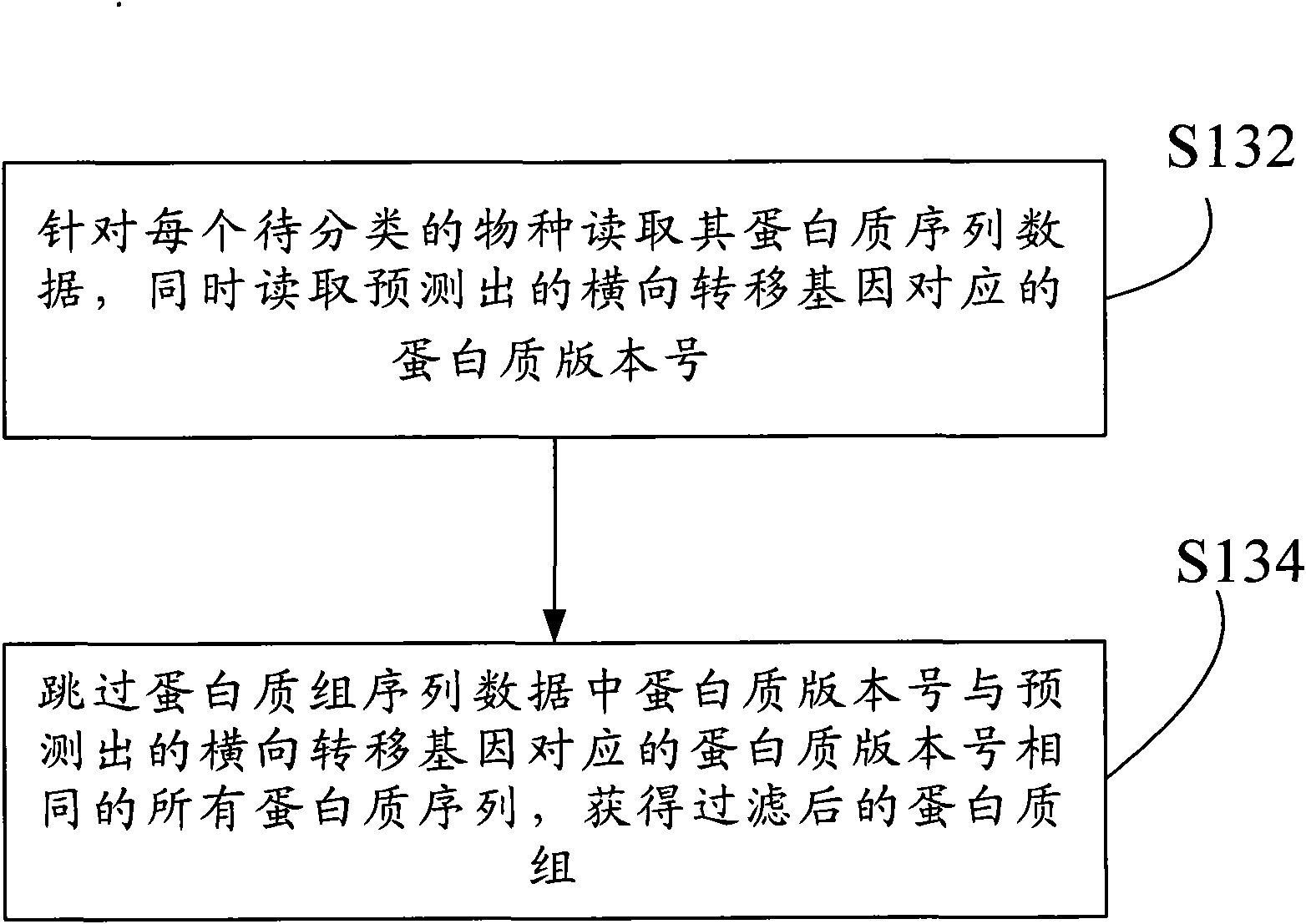
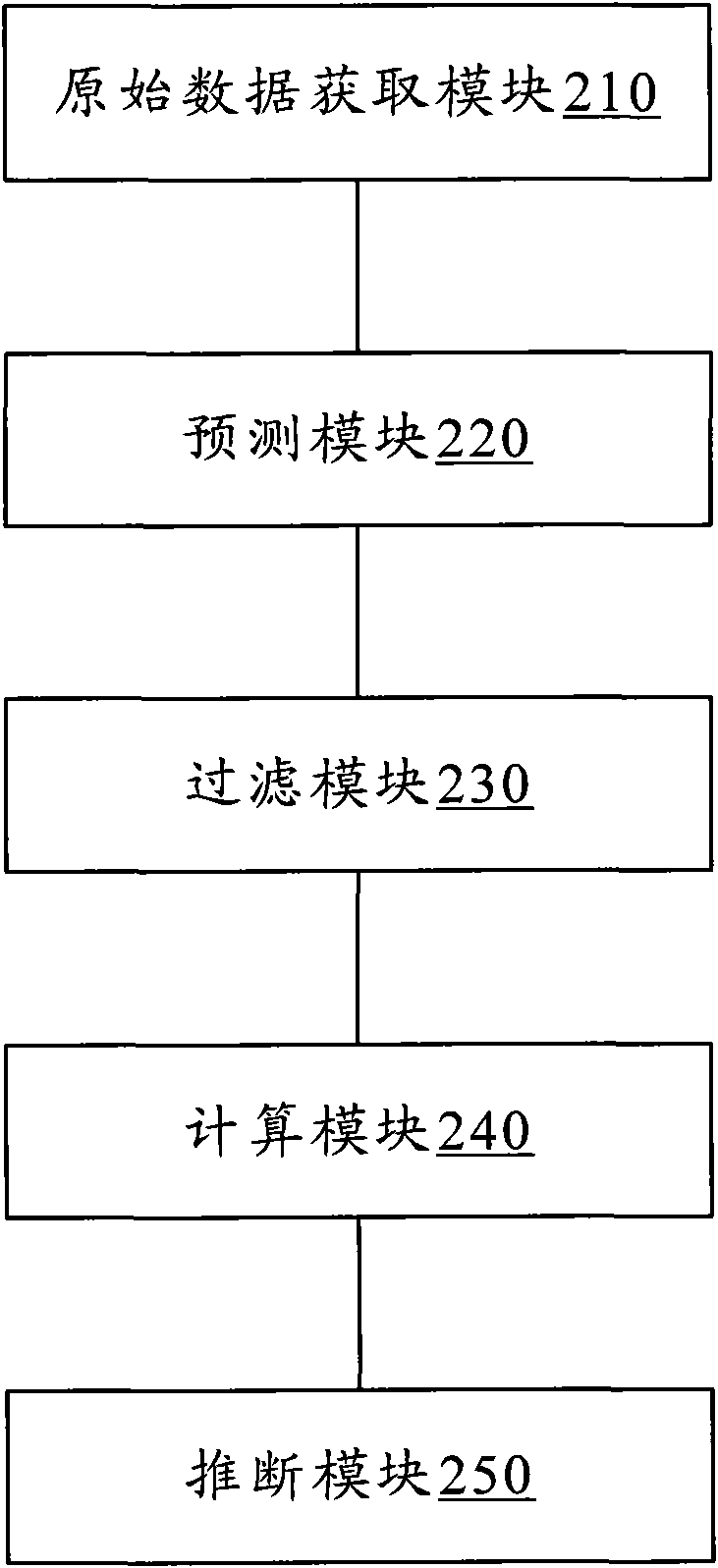
Method for filtering, evolving and classifying proteome and system thereof
InactiveCN101840467AAchieve exclusionReasonable inferenceSpecial data processing applicationsEvolutionary biologyTransfer geneOriginal data
The invention discloses a method for filtering, evolving and classifying proteome and a system thereof. The method comprises the following steps of: an original data acquiring step: acquiring the genome data and the proteome data (S110) of species to be classified; a predicting step: predicting a transversal transfer gene (S120) in a genome sequence based on the genome data; a filtering step: filtering a proteome sequence (S130) corresponding to the transversal transfer gene in the proteome based on the proteome data and the transversal transfer gene; a calculating step: calculating the evolving distance (S140) between the species with the filtered proteome sequence; and an inferring step: inferring a phylogeny relation (S150) between the species by utilizing the evolving distance between the species. According to the invention, existing interference information which is caused by transversal gene transfer during deducing a biogenesis process can be eliminated so that the deduced biogenesis process is more accurate to provide a more reliable classification basis for biological evolution and classification.
Owner:GRADUATE SCHOOL OF THE CHINESE ACAD OF SCI GSCAS
Quick identification of powdery mildew resistant gene for barley
InactiveCN104561032AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementGenetic engineeringResistant genesOrganism
The invention provides quick identification of a powdery mildew resistant gene for barley, relates to subject knowledge of plant comparative genomics, genetics, bioinformatics and the like and belongs to the field of plant biotechnology science. The quick identification mainly comprises the following steps: (1) downloading a barley whole-genome sequence and acquiring an MLO gene; (2) identifying the MLO gene; (3) establishing a phylogenetic relationship of the MLO gene; and (4) comparing an MLO powdery mildew gene. According to the quick identification, the mining period of the barley powdery mildew gene is effectively shortened to facilitate quick identification of the powdery mildew gene; the corresponding coseparation functional markers (SNP, SCAR and the like) are developed through the identified candidate powdery mildew gene and quickly used for molecular marker auxiliary selection of the powdery mildew resistant gene, and the accuracy is high; through combination with other anti-disease gene molecular markers, creation of a multi-resistant breeding material can be carried out, the breeding period is shortened, and the breeding efficiency is improved; and a foundation is laid for stating the barley powdery mildew resistance molecular mechanism.
Owner:CHANGSHU CITY DONGBANG TOWN LIMU VEGETABLE PROFESSION COOP
Microsatellite primer for macrobrachium nipponensis diversity analysis and application thereof
ActiveCN106434974APCR amplification results are stableAmplification results are stableMicrobiological testing/measurementDNA/RNA fragmentationStructure analysisGermplasm
The invention discloses a microsatellite primer for macrobrachium nipponensis diversity analysis. The microsatellite primer for macrobrachium nipponensis diversity analysis comprises eleven pairs of microsatellite primers, which are respectively MN01, MN02, MN03, MN04, MN05, MN06, MN07, MN08, MN09, MN10 and MN11; meanwhile, the invention also provides application of the microsatellite primer in macrobrachium nipponensis population genetic structure analysis through a following method comprising the steps: (a) extracting genome DNA (Deoxyribose Nucleic Acid) to be analyzed; (b) adopting the genome DNA as a template, and adopting the microsatellite primer for carrying out PCR (Polymerase Chain Reaction) molecular marker detection; (c) carrying out genetic structure analysis. The microsatellite primer provided by the invention has the characteristics of stable PCR amplification results, high polymorphism and the like, can be used to utilize molecular markers for carrying out genetic diversity analysis, population genetic structure analysis, genetic map construction, gene mapping, variety identification, germplasm preservation, quantitative trait gene analysis, evolution and phylogenetic relationship research and the like, and has a better application value.
Owner:HEBEI UNIVERSITY
Rapid identification method of powdery mildew-resistant genes of cabbage
InactiveCN104561254AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementPlant peptidesRapid identificationOrganism
The invention provides a rapid identification method of powdery mildew-resistant genes of cabbage, relating to subject knowledge such as comparative genomics of plants, genetics and bioinformatics and belonging to the field of plant biologic technical science. The rapid identification method mainly comprises the following steps: (1) downloading a whole genome sequence of the cabbage, and collecting MLO type genes; (2) identifying the MLO type genes; (3) establishing phylogenetic relationship of the MLO type genes; and (4) comparing MLO type powdery mildew genes. According to the rapid identification method, the mining period of powdery mildew genes of the cabbage is effectively shortened, and the rapid identification of the powdery mildew genes is benefited; by developing corresponding coseparation functional markers (SNP, SCAP and the like) by virtue of identified candidate powdery mildew genes, the molecular marker-assisted selection of the powdery mildew-resistant genes can be rapidly carried out, and the accuracy is high; by combining molecular markers of other disease-resistant genes, multi-resistance breeding materials can be created, the breeding age limit can be shortened, and the breeding efficiency can be improved; a foundation is laid for the exposition of a powdery mildew-resistant molecular mechanism of the cabbage.
Owner:CHANGSHU CITY DONGBANG TOWN LIMU VEGETABLE PROFESSION COOP
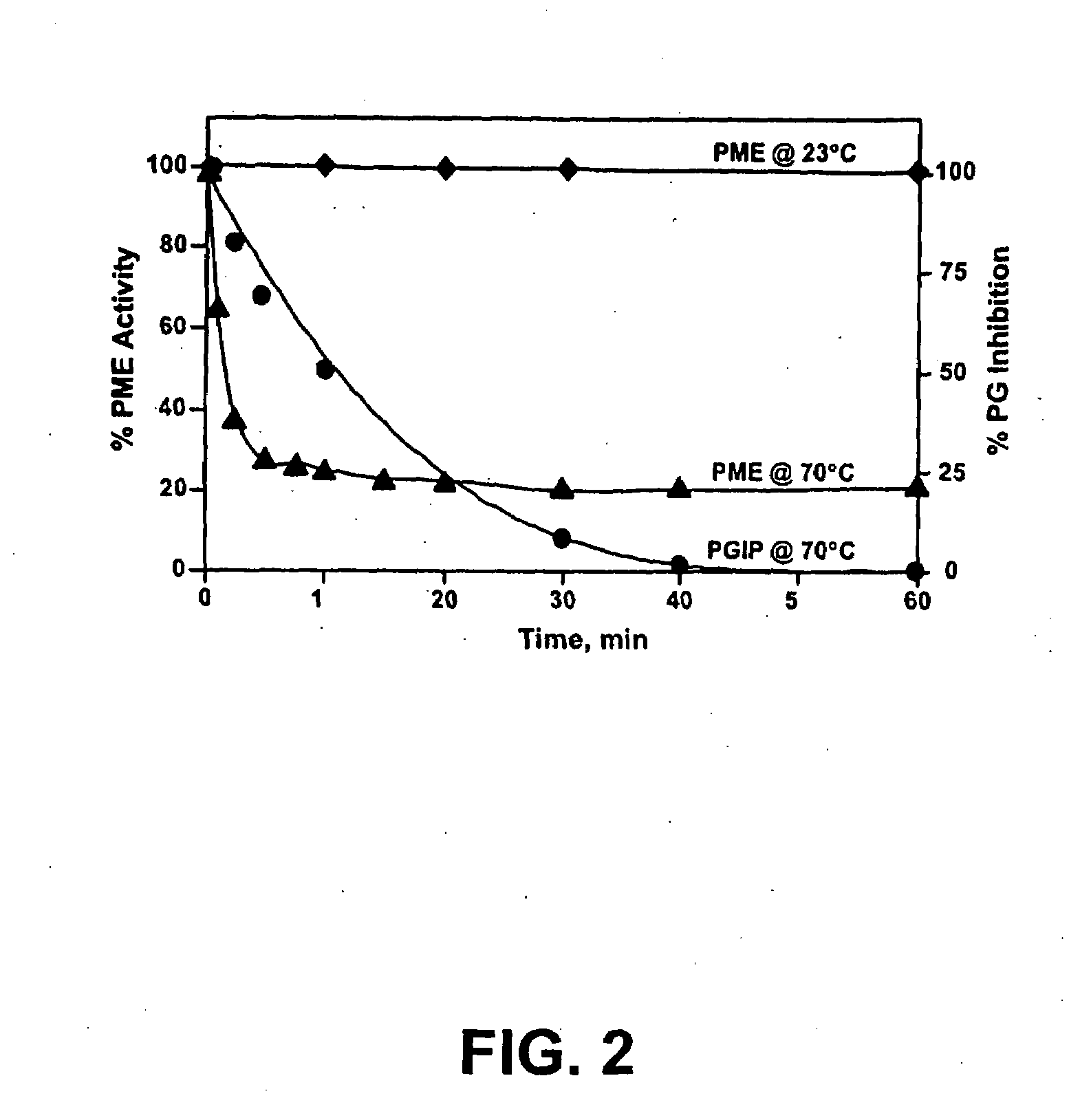
Thermally-tolerant pectin methylesterase
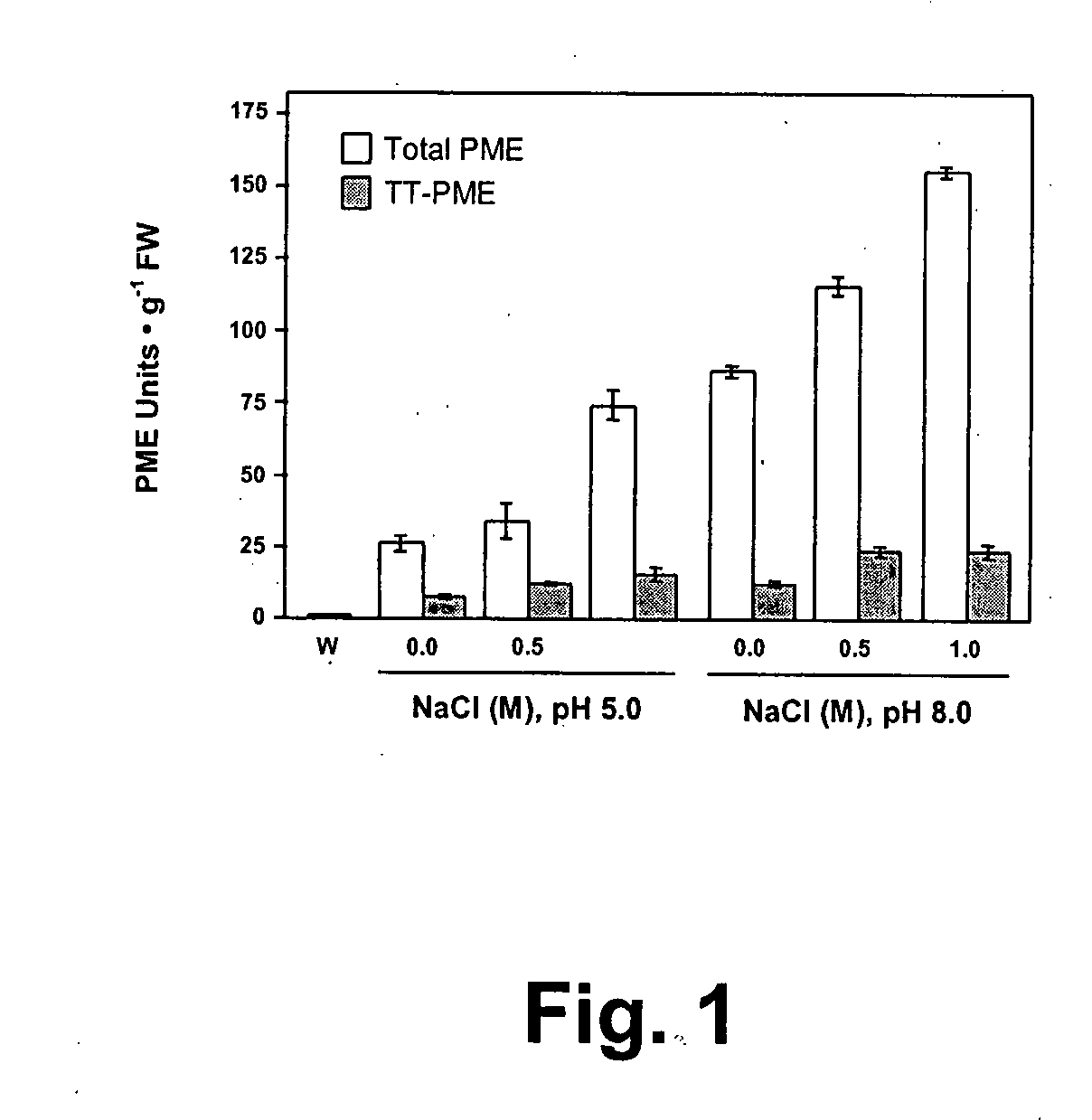
InactiveUS20090130722A1Improve processing and appearanceLow viscositySugar derivativesBacteriaMolecular communicationPlant cell
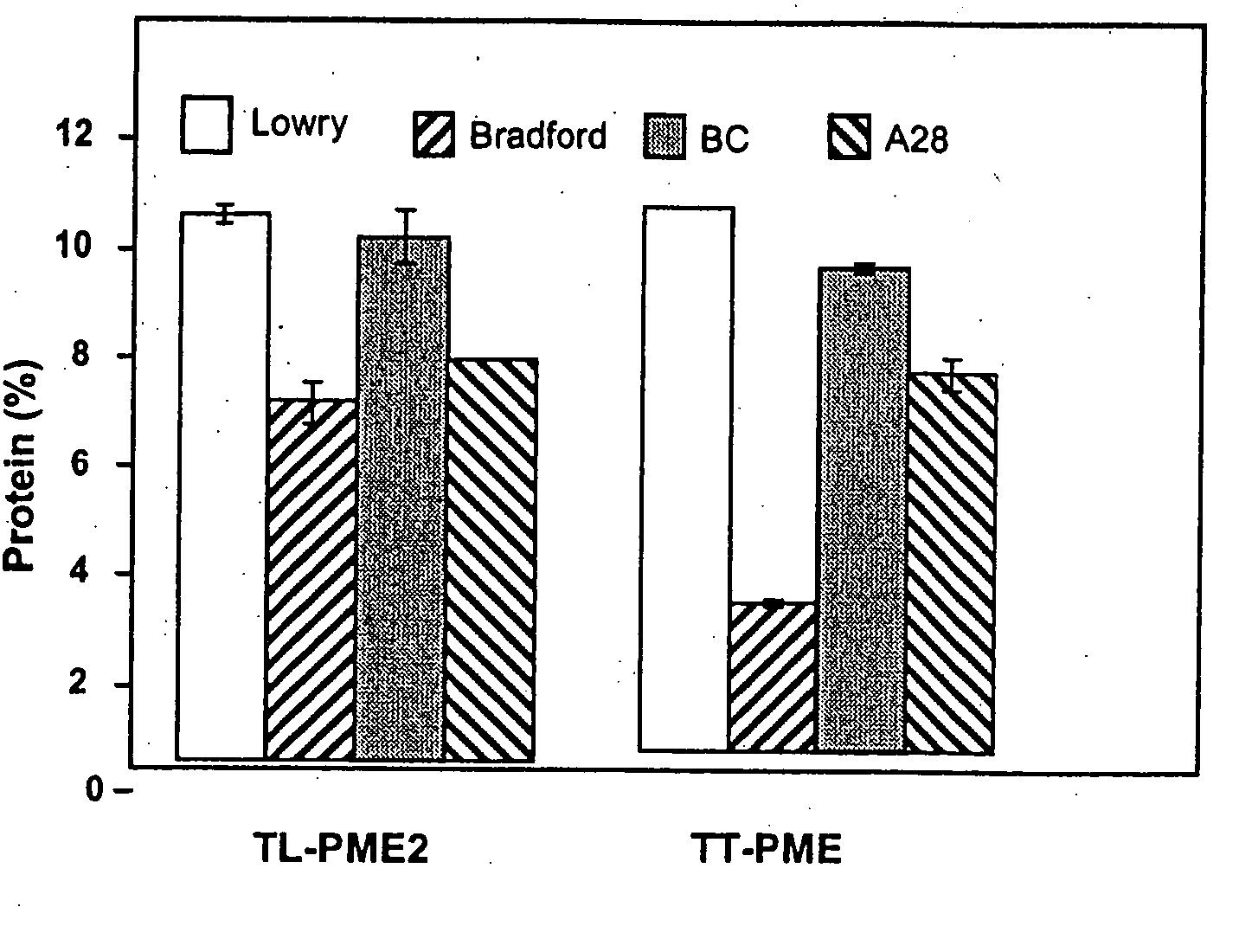
Enzymes accumulated in plant cell walls serve diverse physiological functions including metabolism, polysaccharide structure modification, and molecular communication in interactions with other organisms. Pectin methylesterases are economically important enzymes for their impact on quality and processing properties of fruit and vegetable food products. We have now purified TT-PME to homogeneity from sweet orange finisher pulp and determined the complete corresponding nucleic acid sequence. Purified TT-PME was observed by SDS-PAGE as two doublet bands with molecular masses of approximately 46,000 Da and 56,000 Da. Direct Edman sequencing from these proteins showed a common N-terminal peptide. De novo sequencing of eight TT-PME tryptic peptides determined by MALDI-TOF / TOF mass spectrometry provided additional internal sequences. TT-PME did not correspond to any previously reported Citrus spp. PME sequence. Our results show Citrus TT-PME is a distinctive new isoform with phylogenetic relationship closer to PME isoforms in other species rather than to previously described Citrus PME genes.
Owner:UNITED STATES OF AMERICA
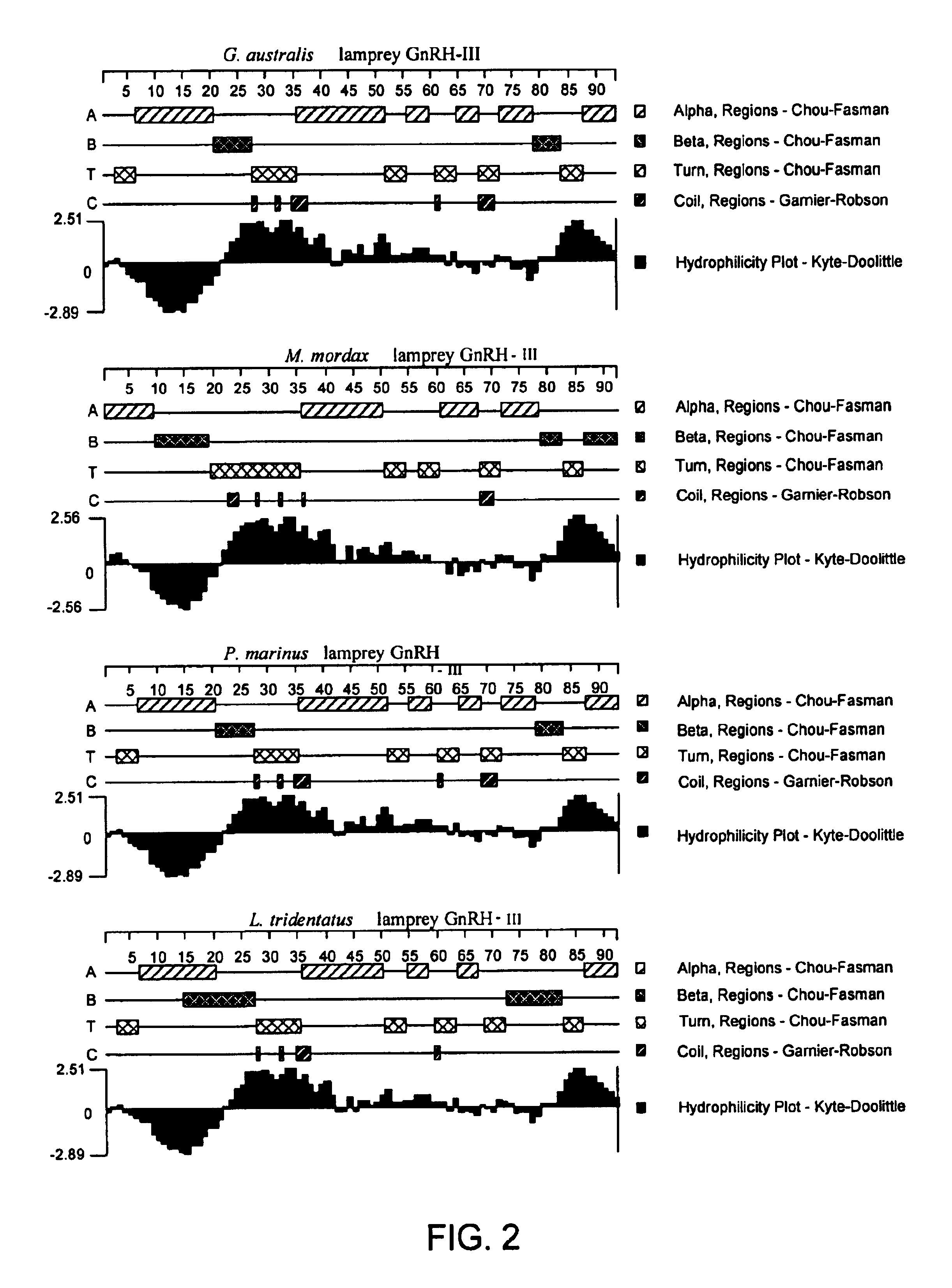
Polynucleotides encoding lamprey GnRH-III
InactiveUS6949365B2The method is simple and safeEasy and less-expensive and saferBacteriaGenetic material ingredientsLampreyNucleotide
Novel isolated cDNA encoding the precursor of the novel gonadotropin-releasing hormones in four species of the fish lamprey is disclosed. The use of such cDNA's in studying the phylogenetic relationship of various species of fish and vertebrates overall; and the use of such cDNA's for controlling the gonadal development and spawning of fish, including inducing and inhibiting maturation, spawning and reproduction, is also described. Methods by which the novel peptides may be administered to fish to control their reproduction are also described.
Owner:NEW HAMPSHIRE UNIV OF
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com