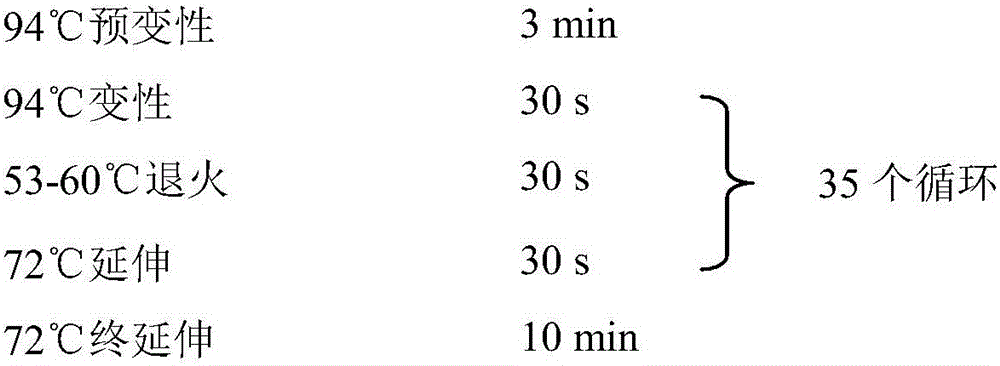Microsatellite primer for macrobrachium nipponensis diversity analysis and application thereof
A diversity analysis and microsatellite technology is applied in the field of microsatellite primers for diversity analysis of Macrobrachium japonicum, and achieves the effects of high polymorphism, good application value and stable PCR amplification results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The acquisition of embodiment 1 microsatellite primer
[0035] Macrobrachium japonica was purchased from the Baiyangdian aquatic product market, and its muscle tissue was taken under sterile conditions, total RNA was extracted from the muscle tissue, and sent to a biological company for transcriptome sequencing to obtain transcriptome data; the assembly length of more than 1kb was analyzed using MISA software unigenes for SSR identification, the identification criteria are: the minimum repeats of precise SSR markers containing two, three, four, five and six nucleotide types are 9, 6, 5, and 4 times respectively, and SSR markers are performed using SSRHunter1.3 Screening to ensure that the front and rear flanks of the sequence are of sufficient length for the design of primers. Use Primer Permier 6 to design primers based on the screened SSRs; the main parameters of the design are: the optimal length of the primer is 18-25 bp, the length of the PCR product fragment is 10...
Embodiment 2
[0039] Embodiment 2 Utilizes microsatellite primers to the analytical method of Macrobrachium japonicus genetic structure
[0040] (1) Genomic DNA extraction: The total genomic DNA of the sample to be analyzed was extracted by referring to the phenol-chloroform method provided in the "Molecular Cloning Experiment Guide" (Sambrook & Russell, 2001). DNA quality inspection: take 2 μL of total genomic DNA and run it on an agarose gel with a mass ratio concentration of 1.5%, and observe whether the DNA is degraded and whether there are protein residues after EB staining and ultraviolet gel imaging system imaging; in addition, the extracted The concentration of the DNA sample was measured with a NanoDrop 2000 ultra-micro spectrophotometer, and the total DNA sample of the genome was uniformly diluted to 50 ng / μL with the measured DNA concentration as a reference.
[0041] (2) Use the microsatellite primers to detect molecular markers: use the uniformly diluted genomic total DNA obtaine...
Embodiment 3
[0047] Example 3 Application of Macrobrachium japonicus Genetic Structure Analysis
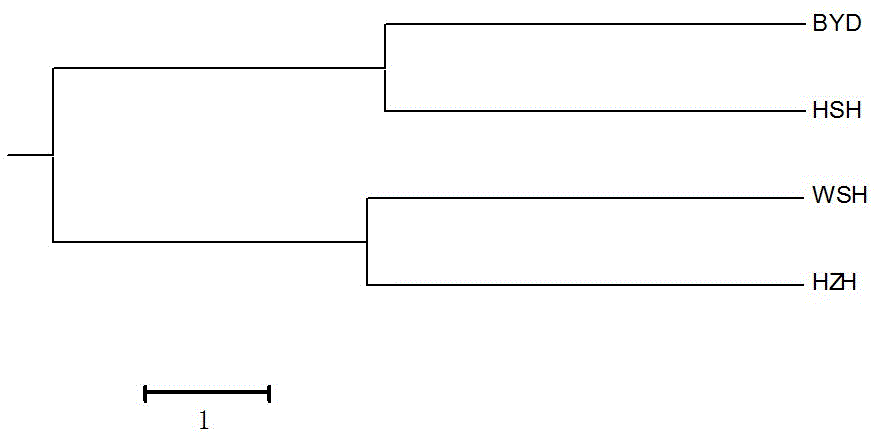
[0048] The analysis population comes from four populations of Macrobrachium japonicus from Hebei Baiyangdian (BYD), Hebei Hengshui Lake (HSH), Shandong Weishan Lake (WSH), and Jiangsu Hongze Lake (HZH). Each population takes 24 samples, a total of 96 Sample, extract the total genomic DNA of each sample according to (1) in Example 2, take the total genomic DNA as the amplification template, and carry out PCR typing detection with 11 pairs of microsatellite primers obtained in Example 1, the amplification system, Amplification conditions and specific amplification steps are the same as step (2) of Example 2, and the amplified products are separated by polyacrylamide gel with a mass ratio concentration of 10%, stained with silver staining, record the amplification results, and detect The fragment size of the amplified product. Some of the amplified fragments are as SEQ ID NO:23 (amplified by MN0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com