Tracing method of cronobacter spp. in infant formula milk powder
A technology of Cronobacter and infant formula, which is applied in the field of traceability of Cronobacter in infant formula milk powder, can solve problems such as restricting the healthy development of the PIF industry, inability to prevent and control Cronobacter, and the impact of pathogenic bacteria
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Activation, purification and identification of embodiment 1 Cronobacter
[0034] The tested strains were 80 strains of Cronobacter, of which 43 were isolated from PIF, 27 were isolated from the processing environment of PIF, and 4 were reference strains of Cronobacter (C. sakazakii ATCC BAA-894, C. sakazakii ATCC 29004 , C.sakazakii ATCC 29544 and C.sakazakii ATCC 12868), 6 strains were Cronobacter type strains (C.malonaticus CDC105877T, C.dublinensis LMG23823T, C.turicensis LMG23827T, C.universalis NCTC9529T, C.condimenti LMGT and C. .muytjensii ATCC 51329T). In addition, E.aerogenes ATCC 13048, E.cloacae ATCC 35030, Escherichia CMCCB 44113 and Escherichia O157 were used as external reference strains. The specific conditions are shown in Table 4, and the above-mentioned tested strains were all from Laboratory 503, Key Laboratory of Dairy Science, Ministry of Education, Northeast Agricultural University.
[0035] Table 4 Tested strains and their sources
[0036] Tabl...
Embodiment 2
[0050] The multi-locus sequence typing of embodiment 2 Cronobacter
[0051] (1) 7 pairs of housekeeping genes specific PCR amplification and gene sequencing
[0052] (1) Specific amplification of 7 pairs of housekeeping genes
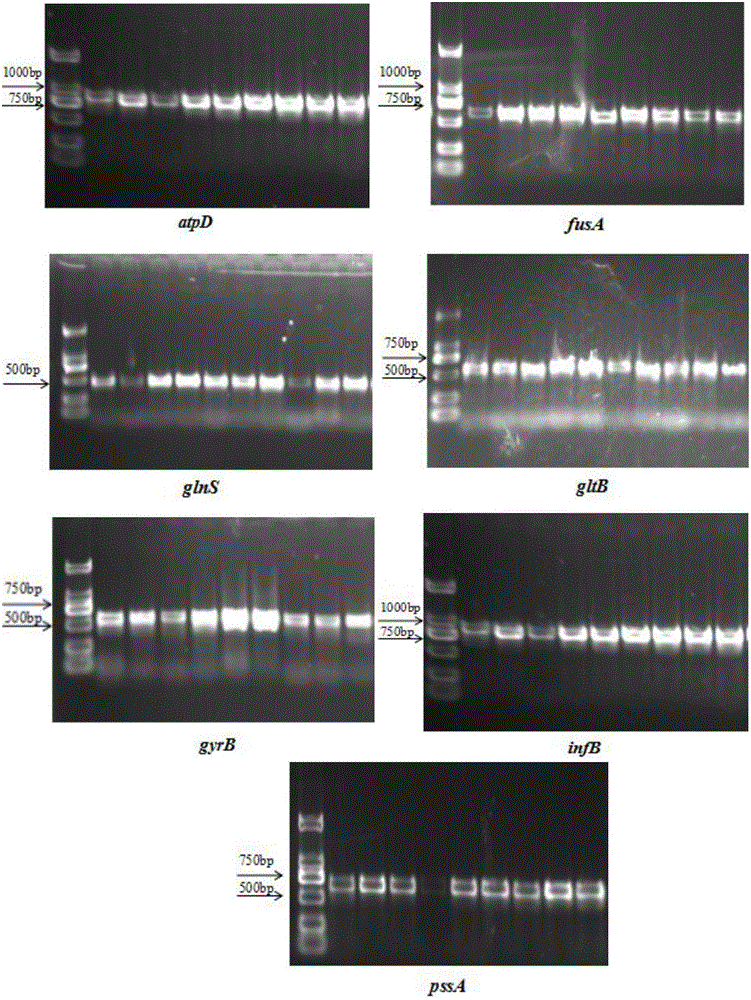
[0053] Synthesize 7 pairs of primers for housekeeping genes (atpD, fusA, glnS, gltB, gyrB, infB, and ppsA) (see Table 3 for the gene sequence), use Cronobacter DNA as a template for specific amplification, and PCR of 7 pairs of primers The amplification system and conditions are the same, as shown in Table 6. The PCR amplification product was checked by 1% agarose gel electrophoresis, and the results were observed and recorded under the UVP ultraviolet gel phase system. The results are as follows: figure 1 shown. The target bands of the 7 housekeeping genes were clear, and the size of the product sequence was judged to be the target bands of the 7 housekeeping genes according to the position of the bands.
[0054] Table 6 7 pairs of housekeeping gen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com