Patents
Literature
60 results about "Gene mining" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The principle reason for gene mining is to identify and isolate genes that are. characterised for conferring essential traits. The widespread use and availability of. molecular biological techniques have allowed for the rapid development and.
Gene mining system and method
InactiveUS6928368B1Cell receptors/surface-antigens/surface-determinantsSugar derivativesBioinformaticsGene mining
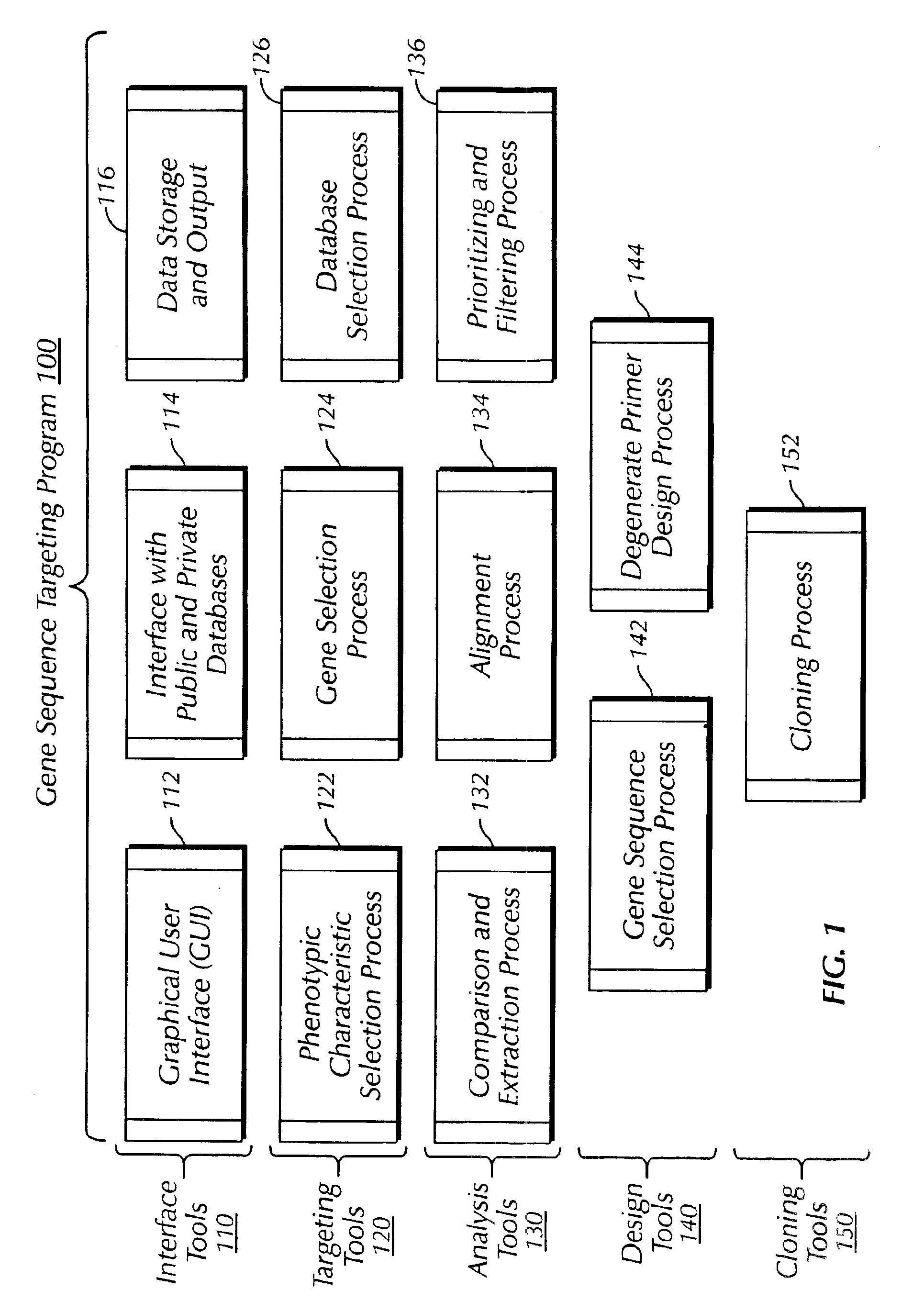
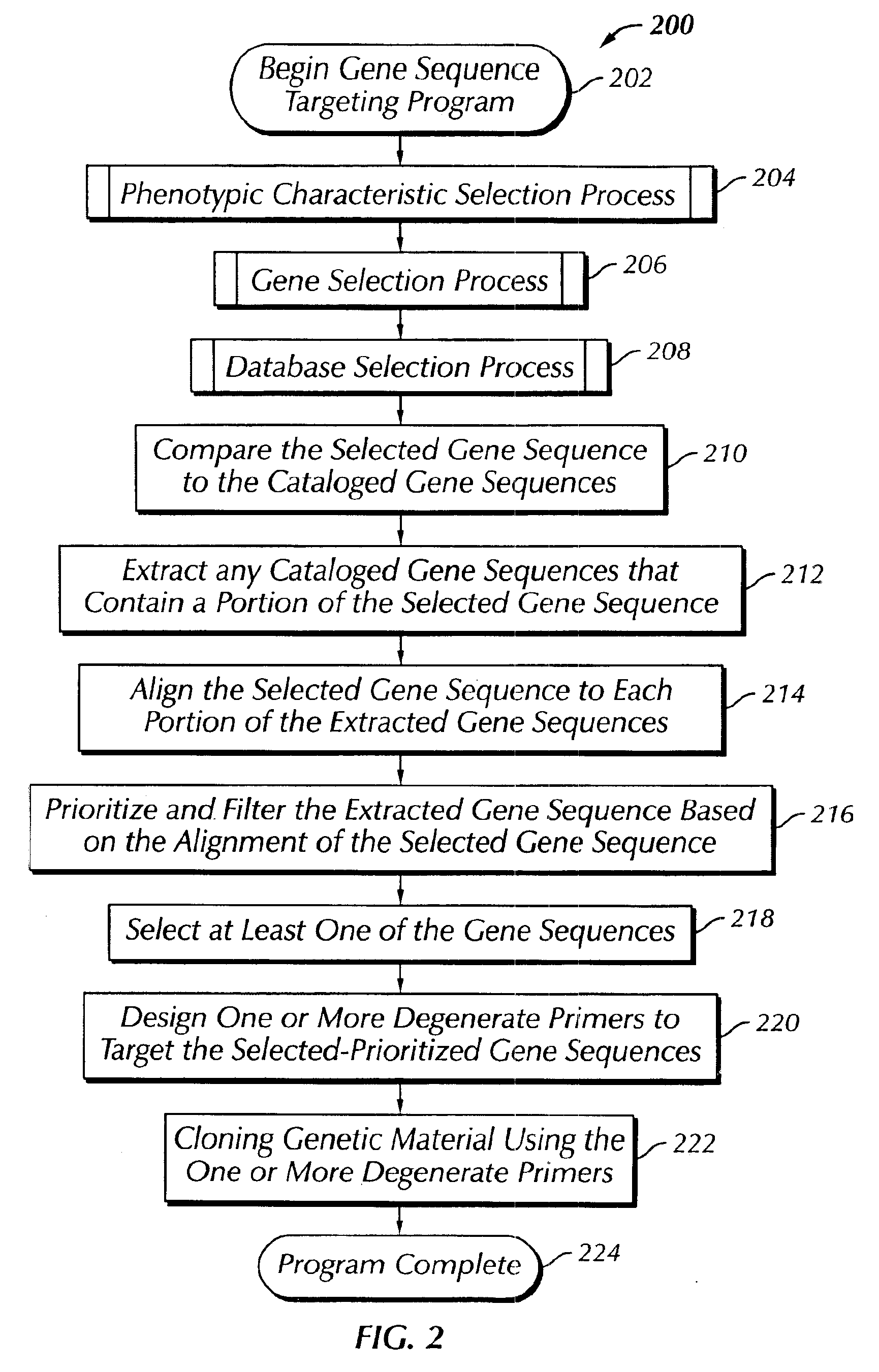
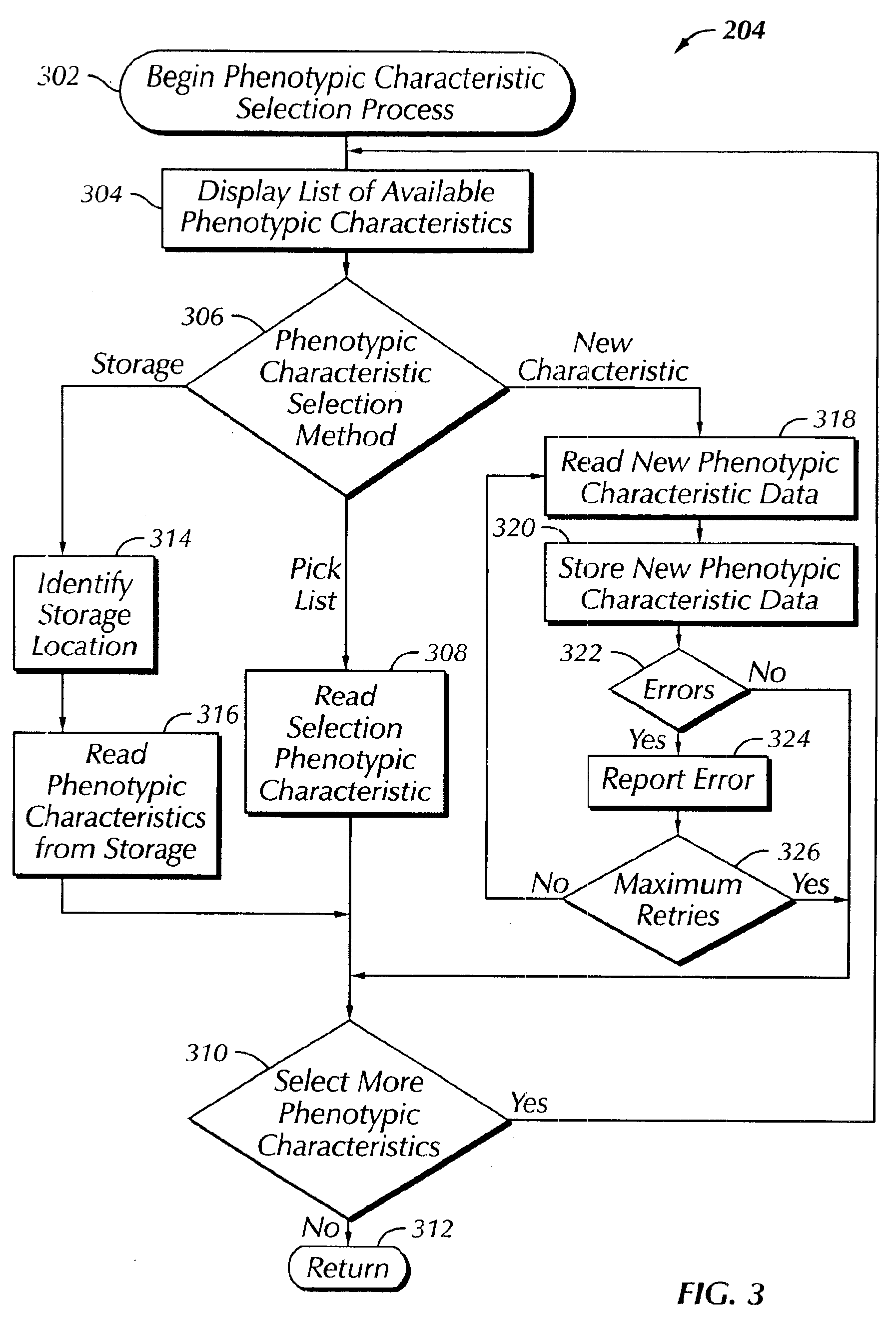
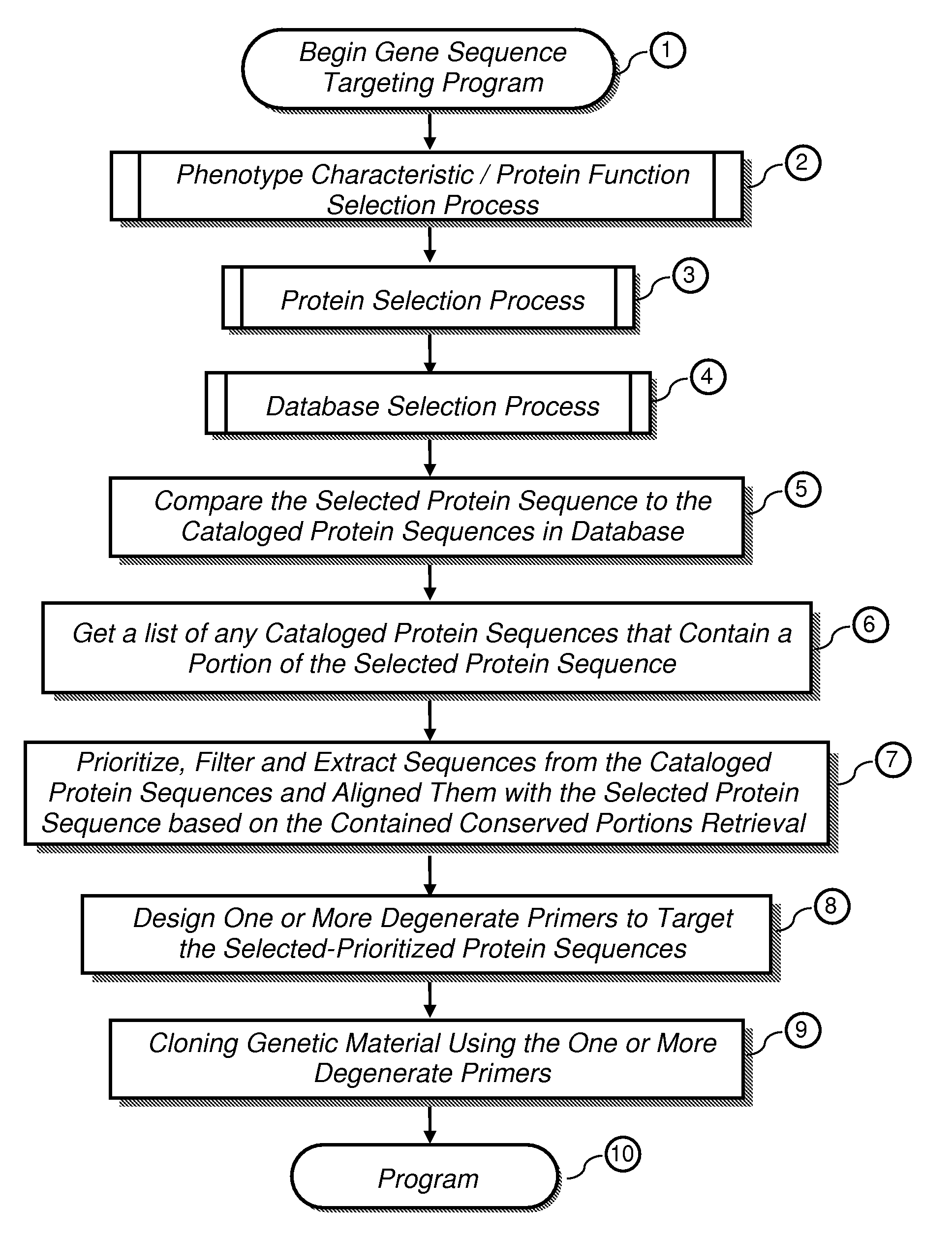
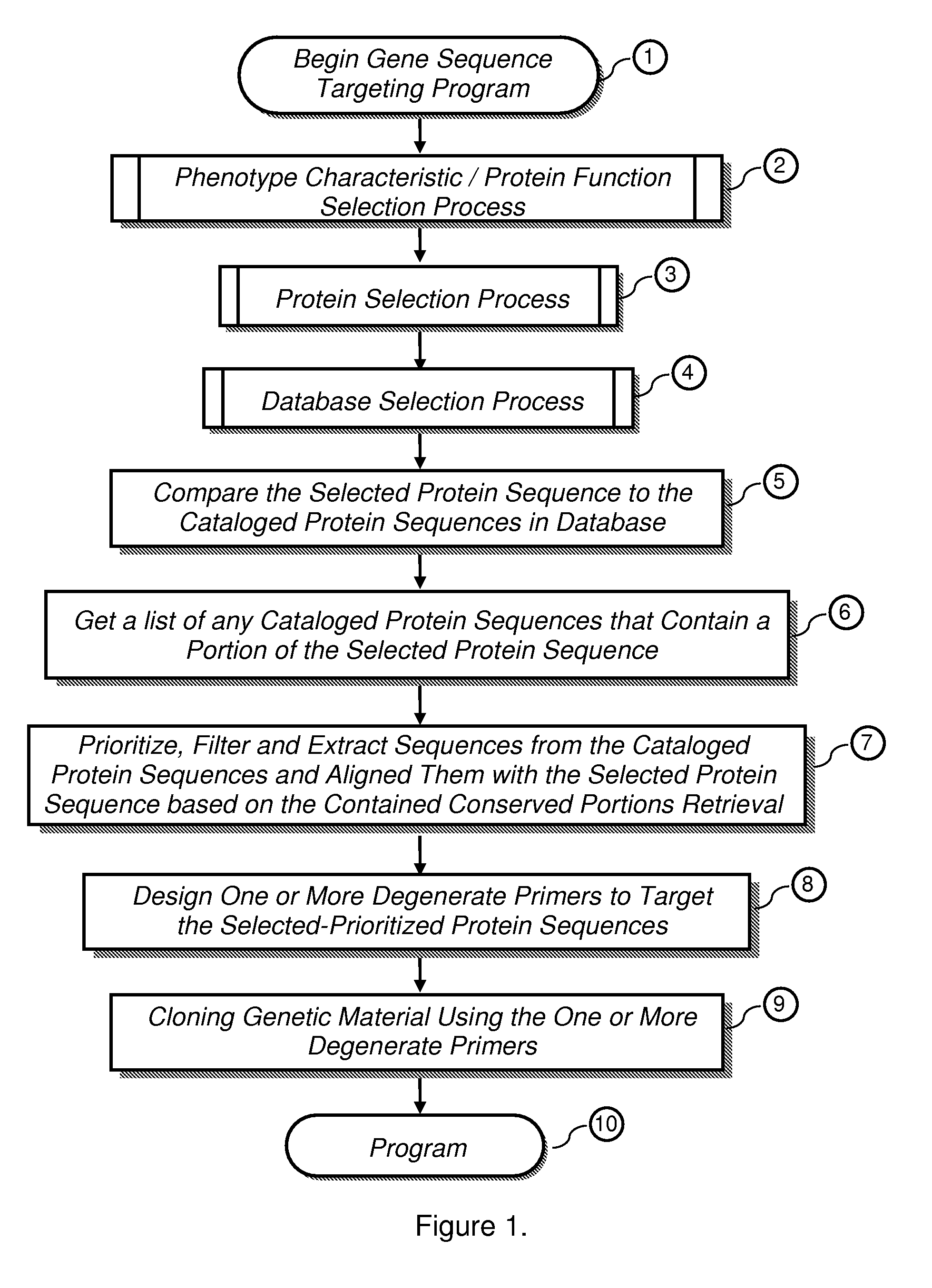
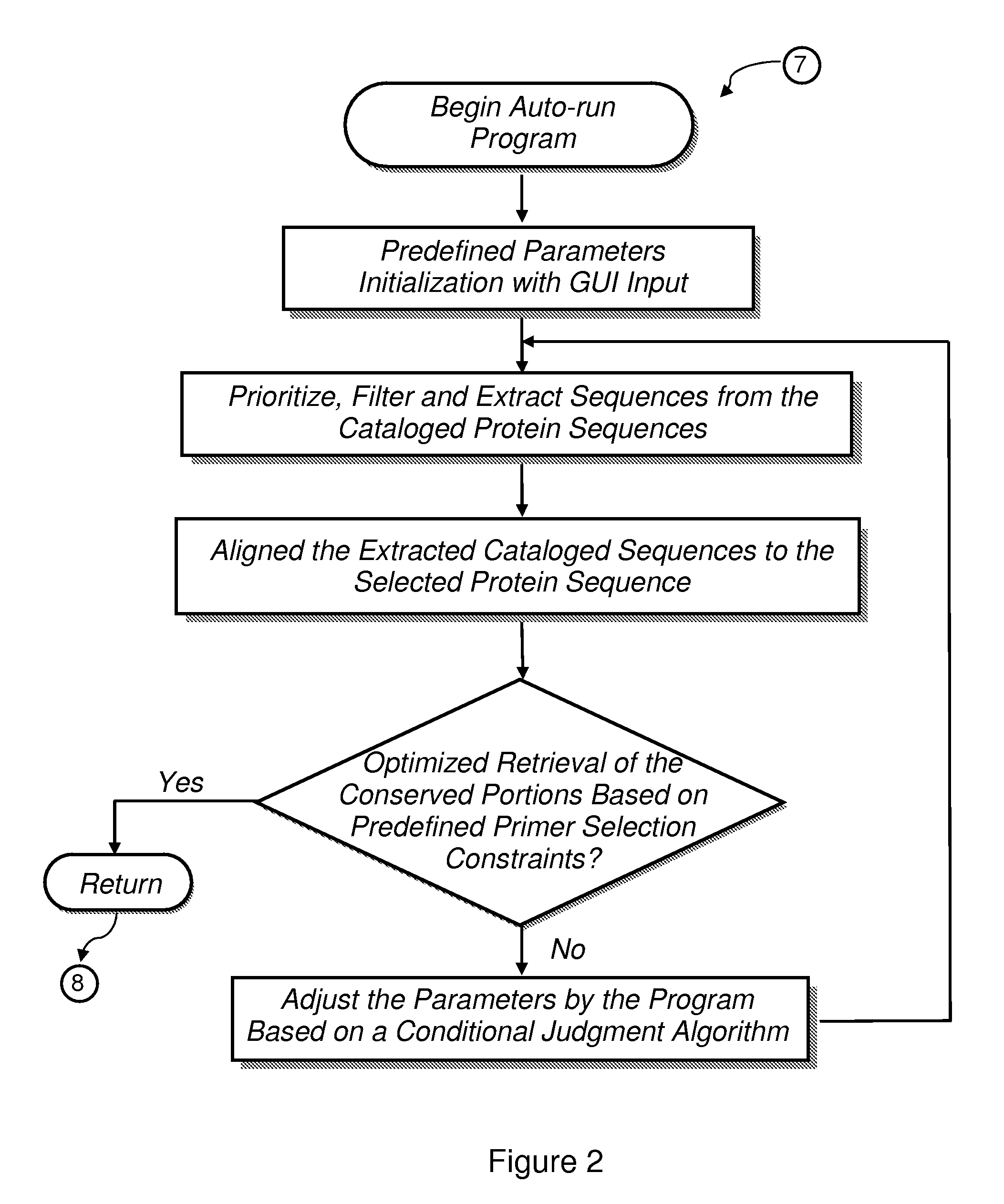
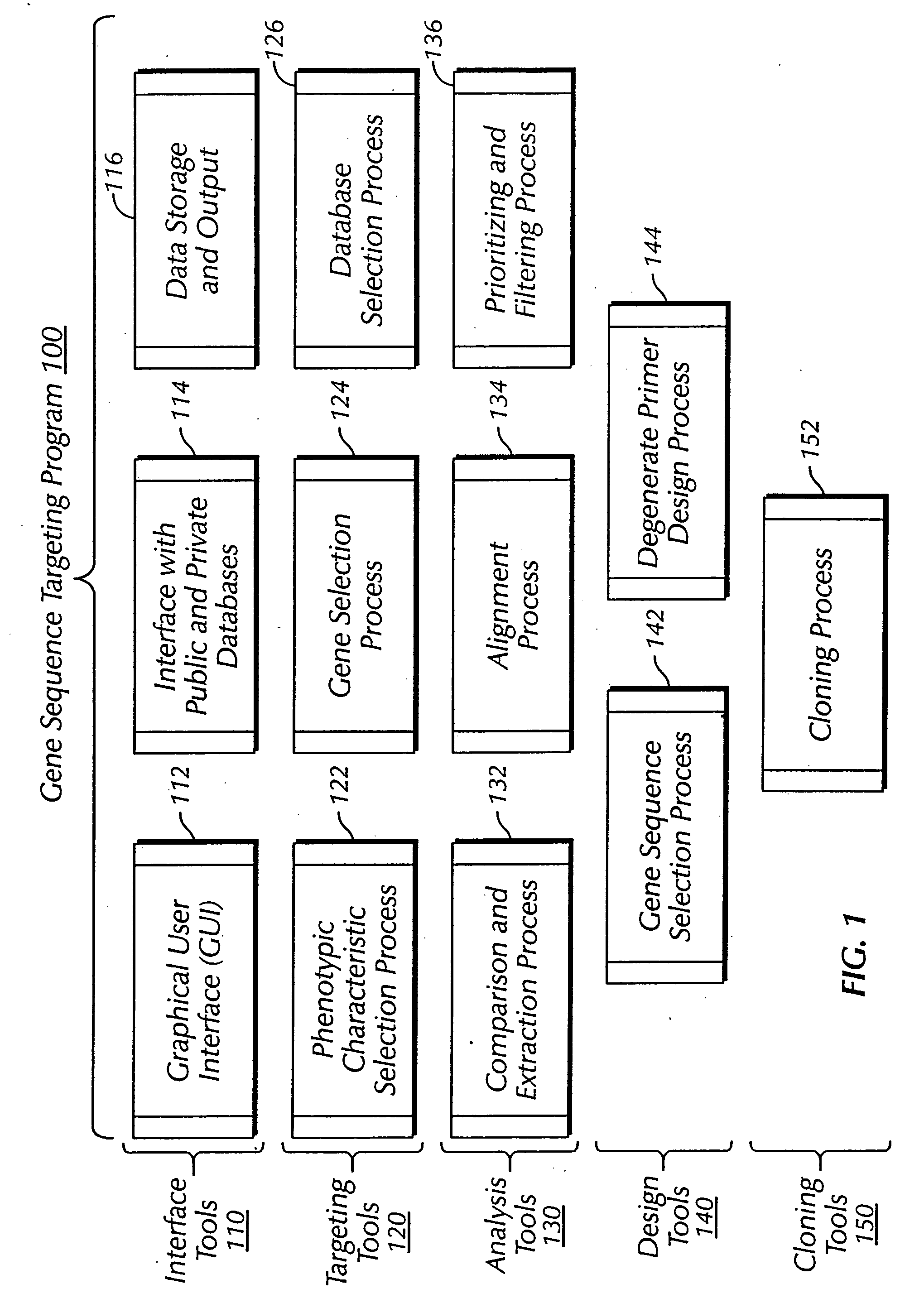
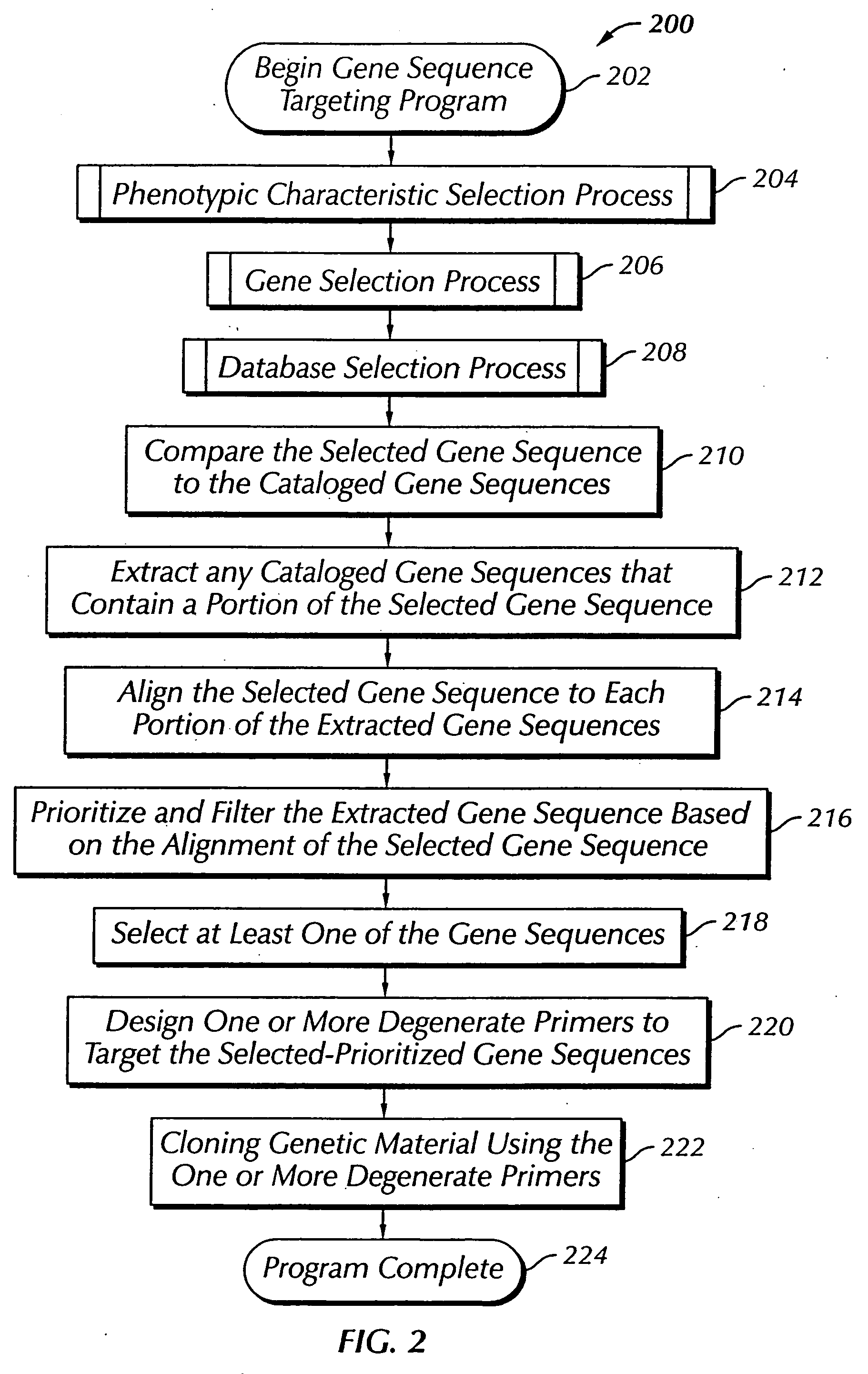
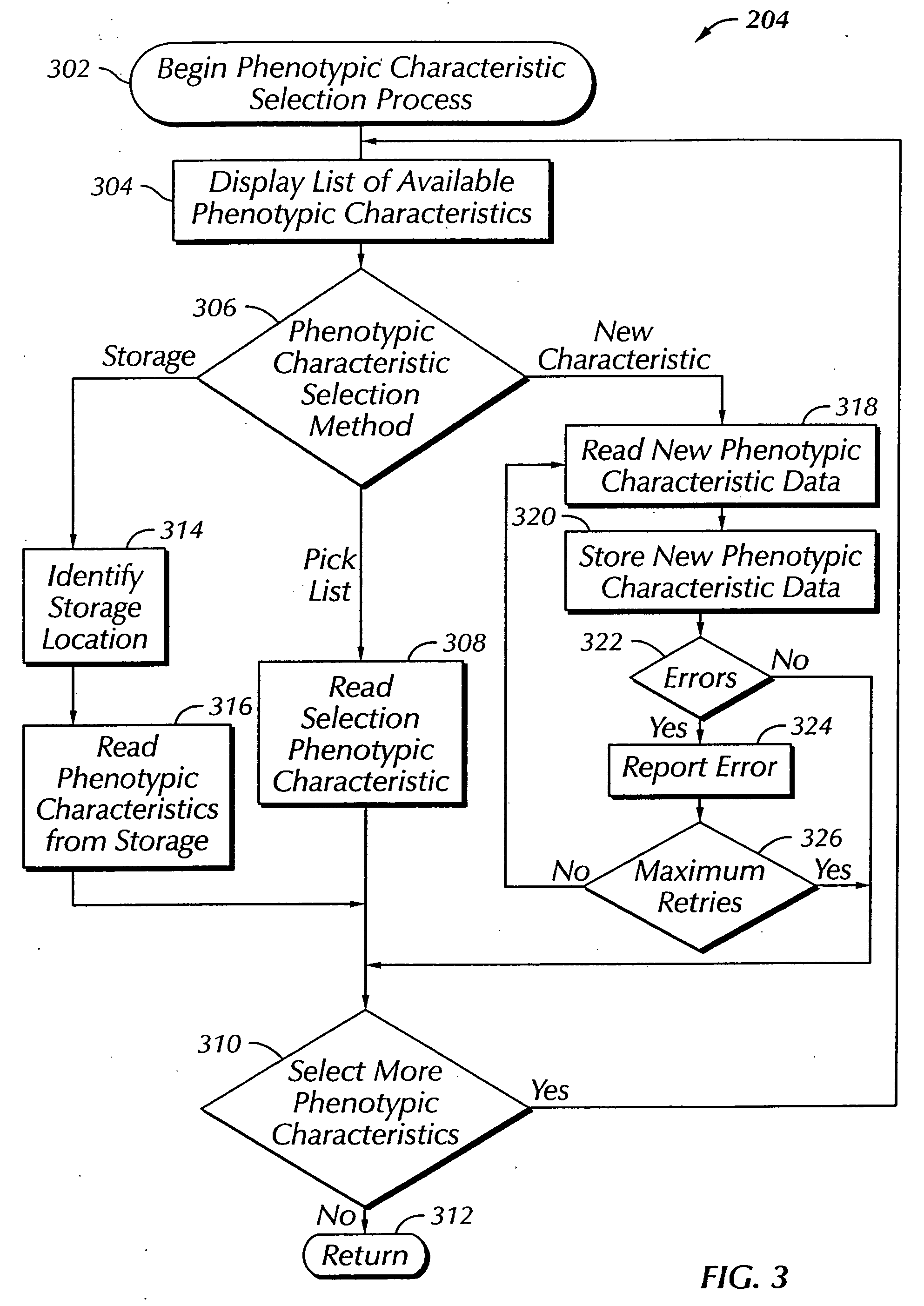
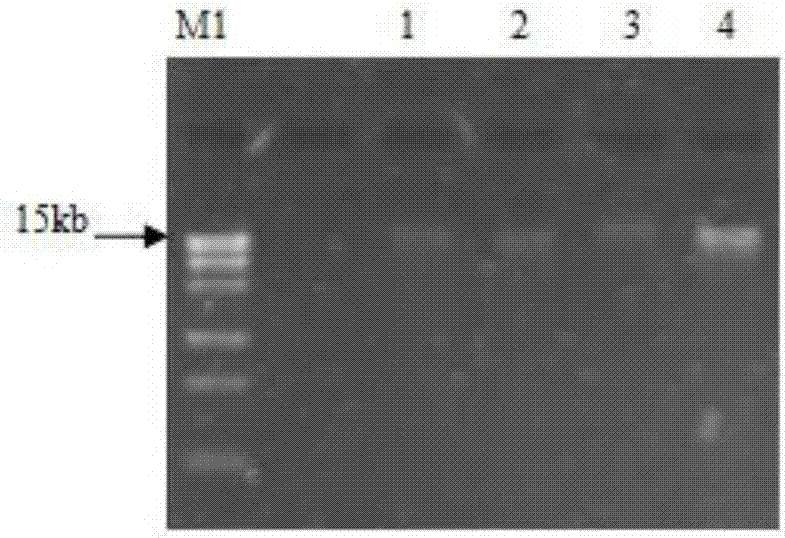
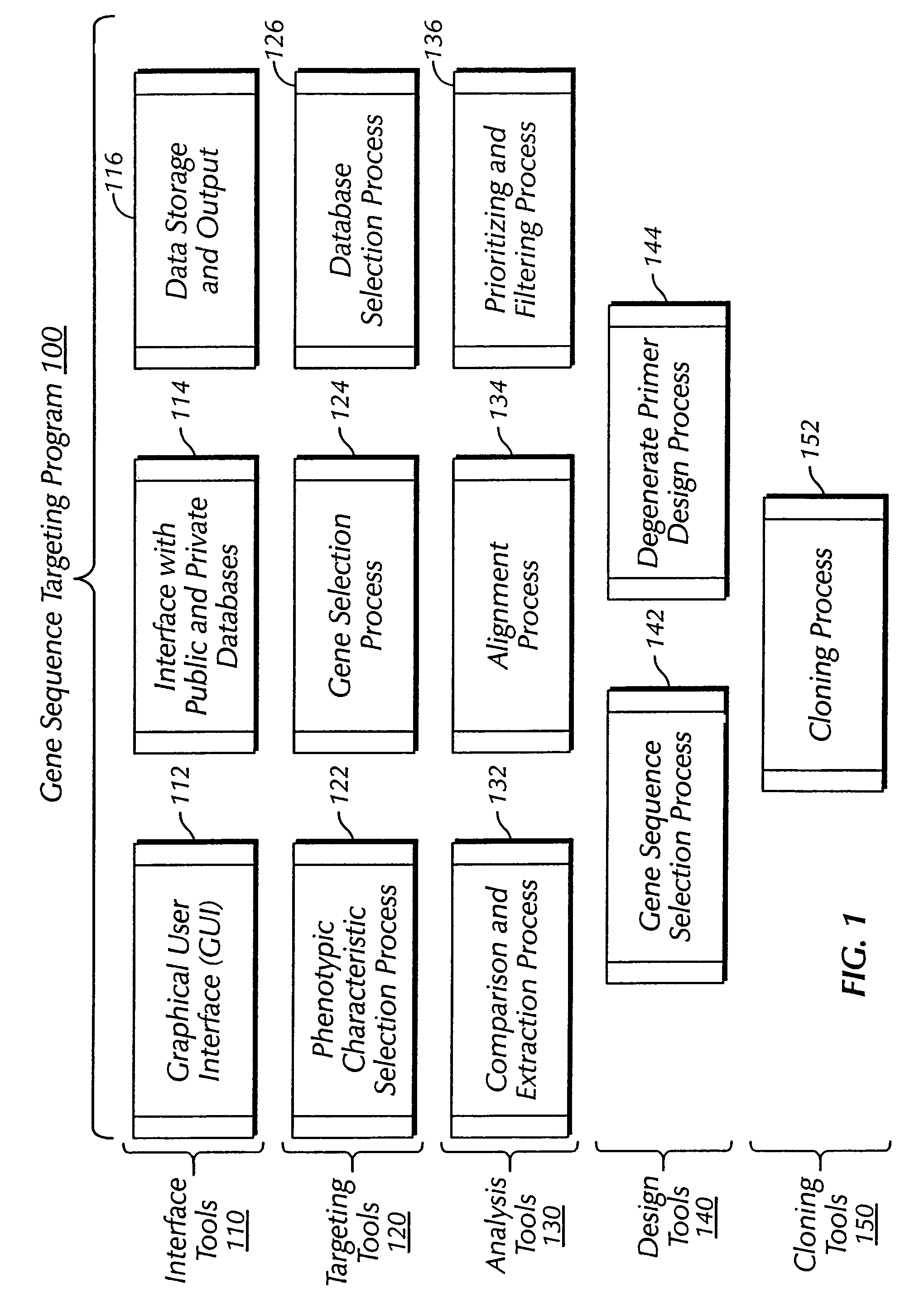
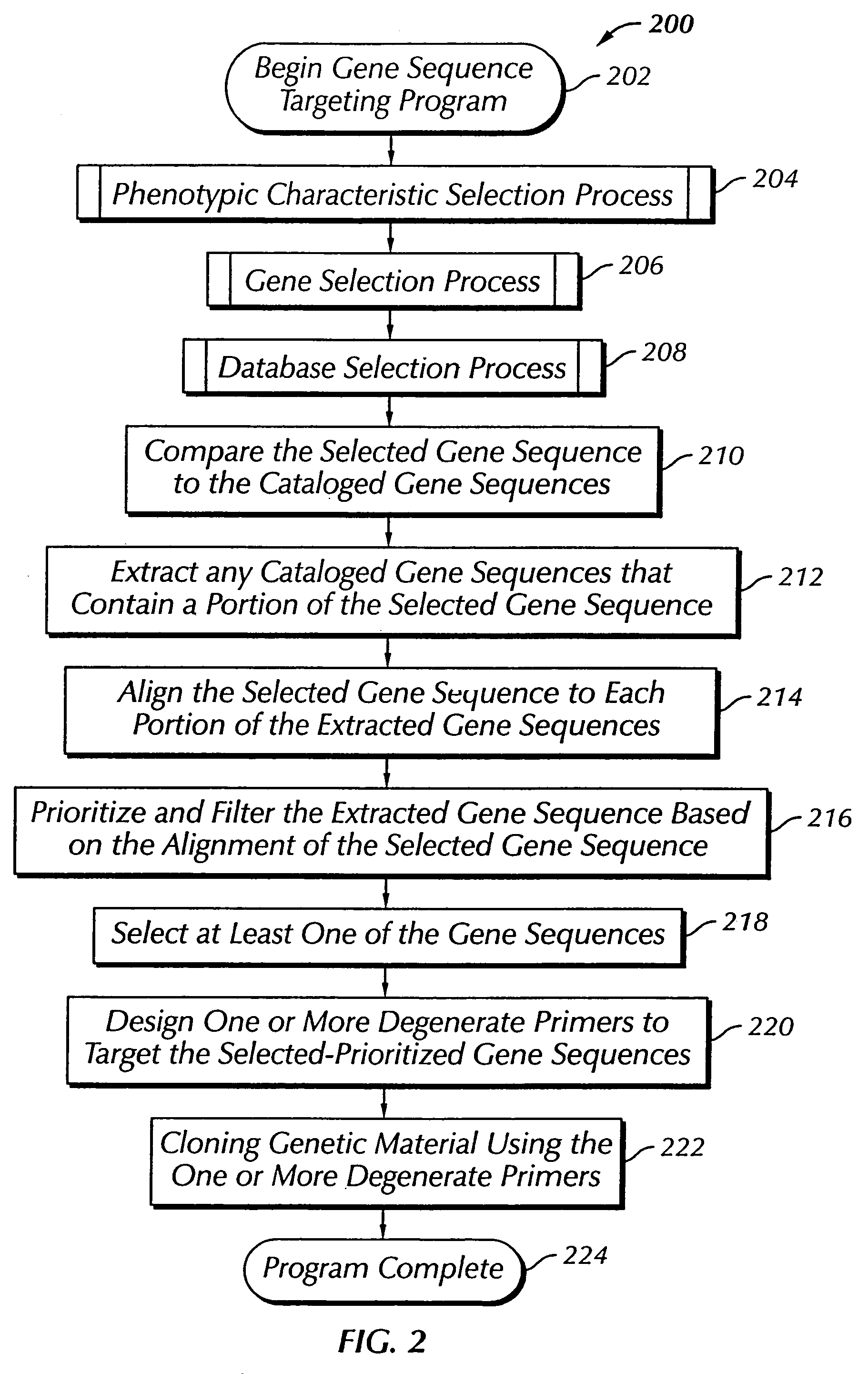
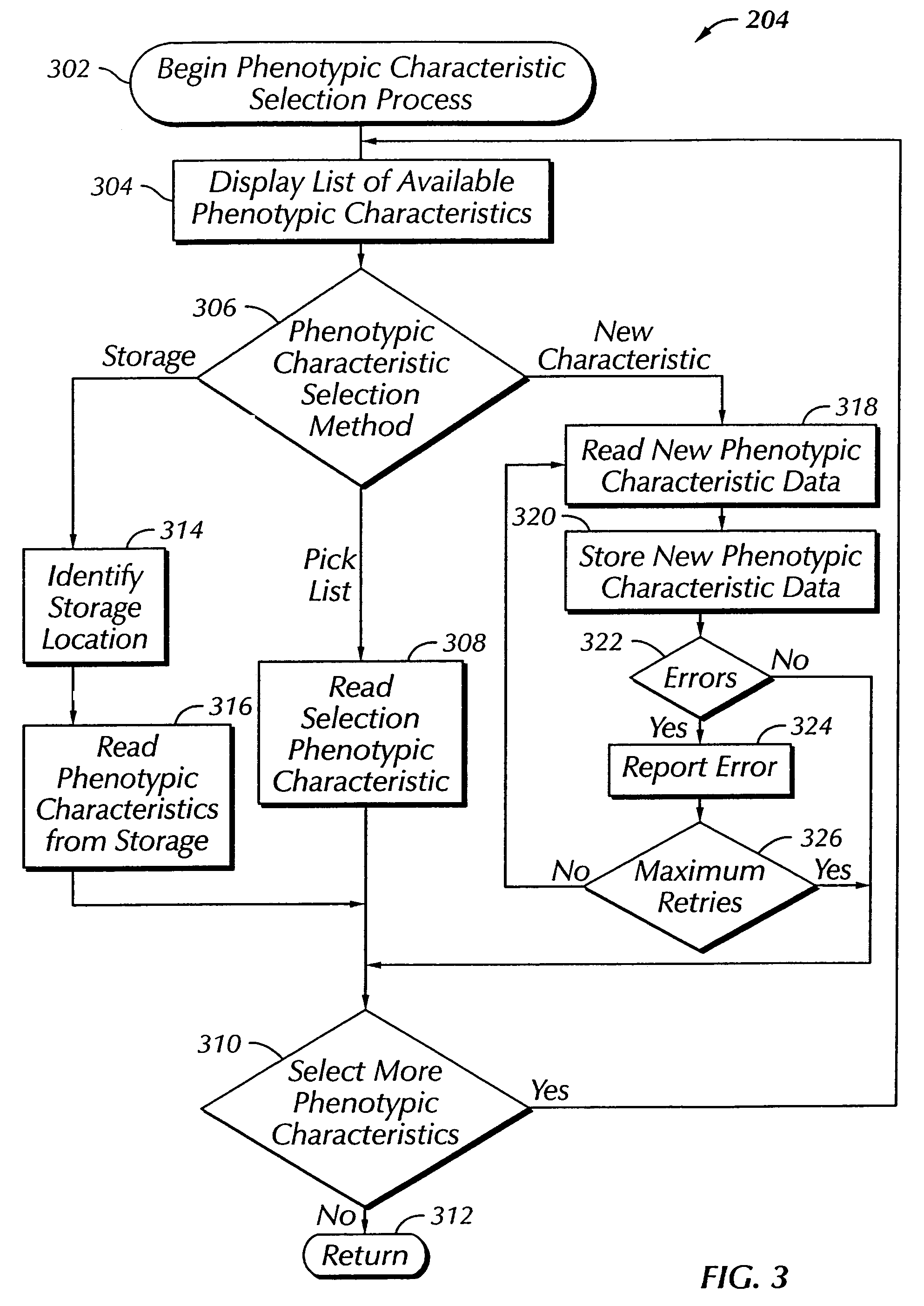
The present invention provides a system, method and apparatus for targeting gene sequences having one or more phenotypic characteristics using a computer. One or more phenotypic characteristics are selected. A gene sequence is then selected that is known to have the selected phenotypic characteristics. In addition, one or more databases containing cataloged gene sequences are selected. The selected gene sequence is compared to the cataloged gene sequences, and any cataloged gene sequences that contain a portion of the selected gene sequence are extracted. The selected gene sequence is aligned to each portion of the extracted gene sequence and the extracted gene sequences are prioritized based on the alignment of the selected gene sequence. At least one of the prioritized gene sequences is selected based on one or more phenotypic criteria. Finally, one or more degenerate primers are designed to target the selected-prioritized gene sequences.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Rapid identification of powdery mildew resistance Lotus corniculatus genes utilizing comparative genomics
InactiveCN102703463AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesBiotechnologyGenetics genomics
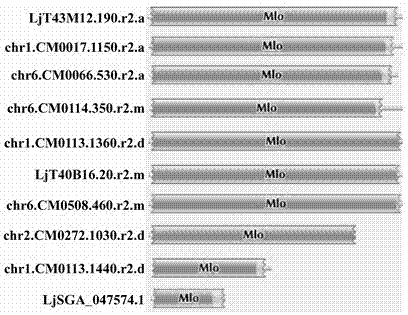
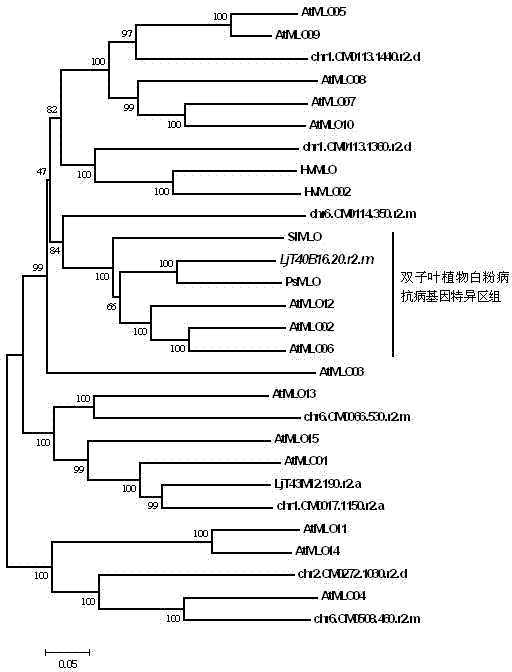
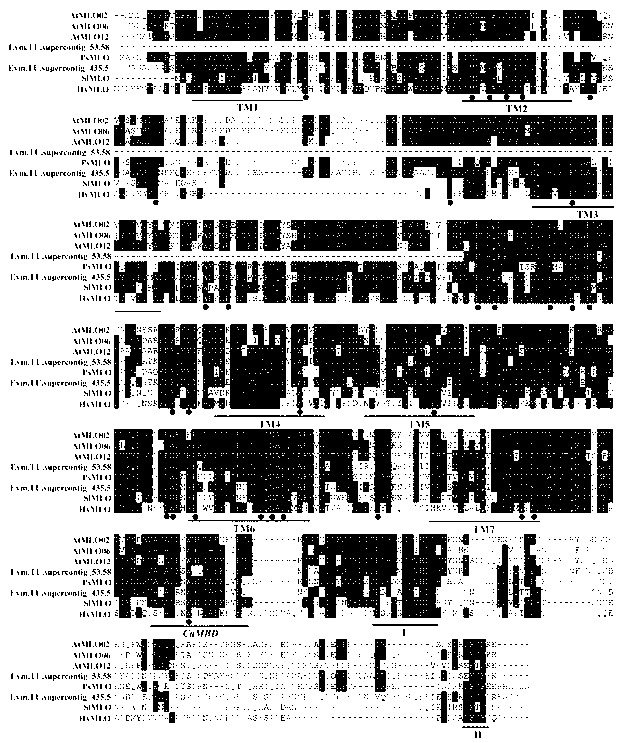
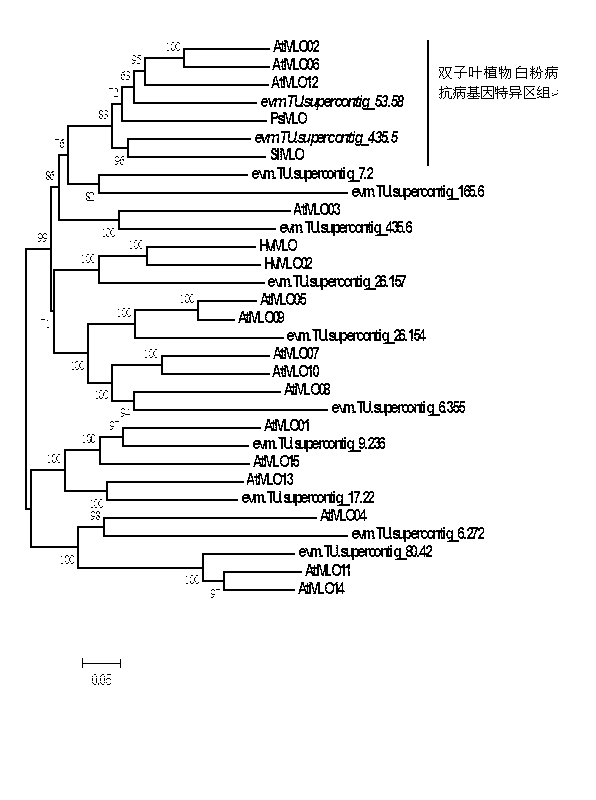
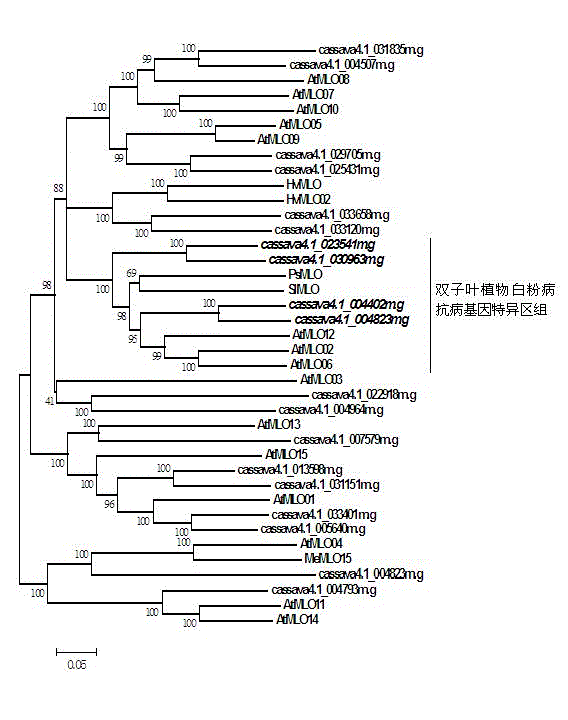
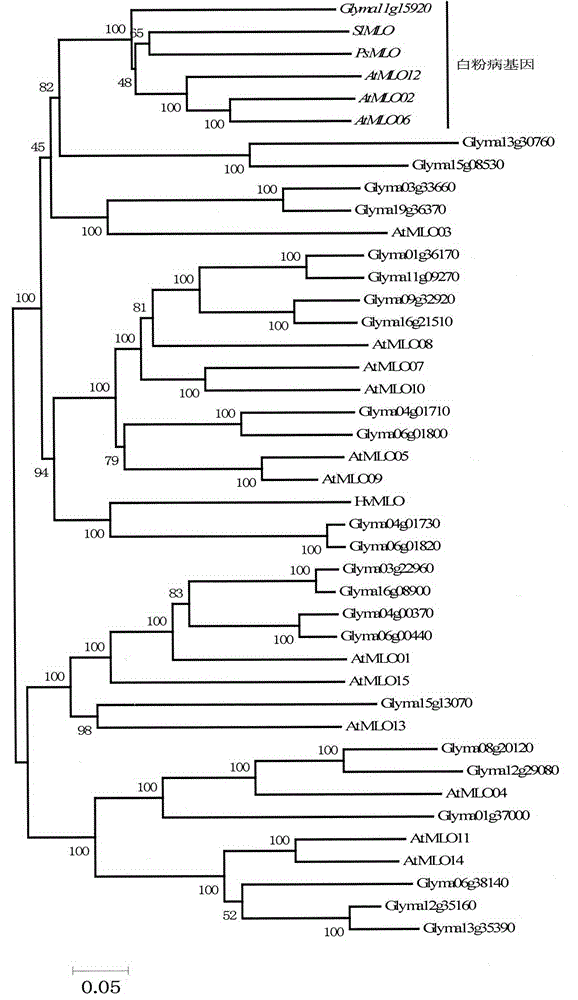
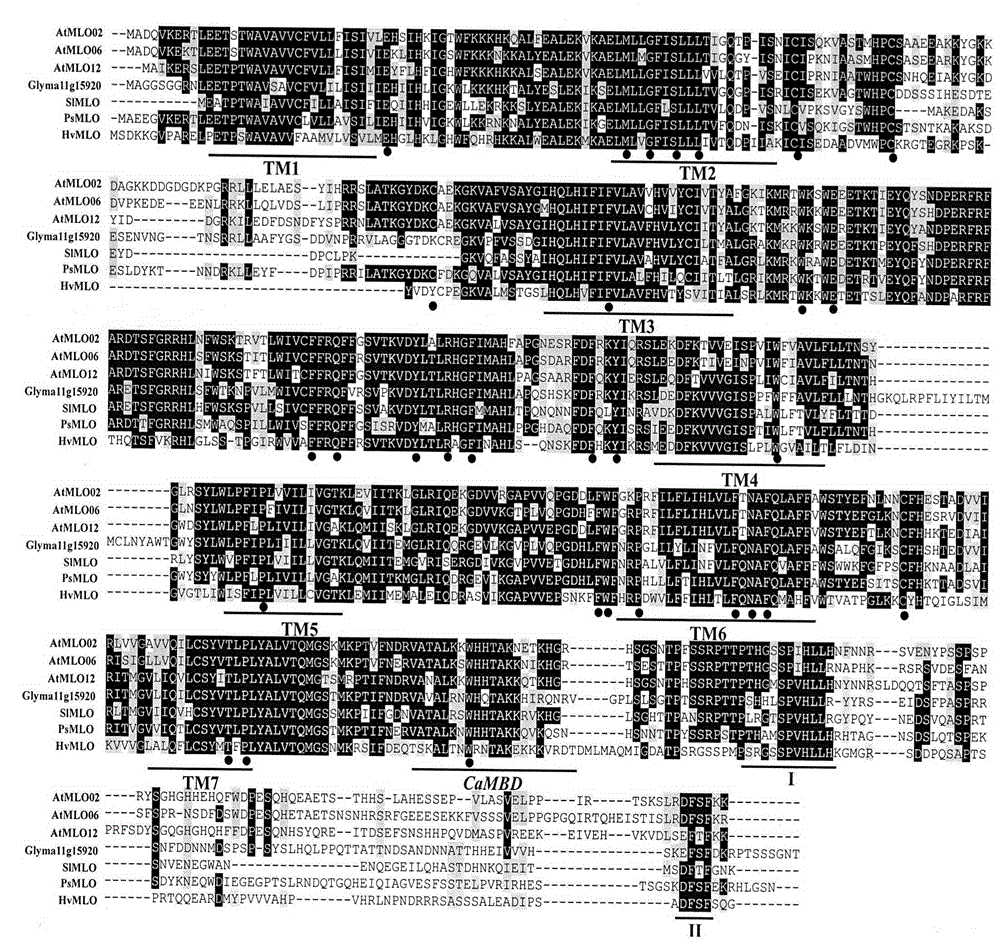
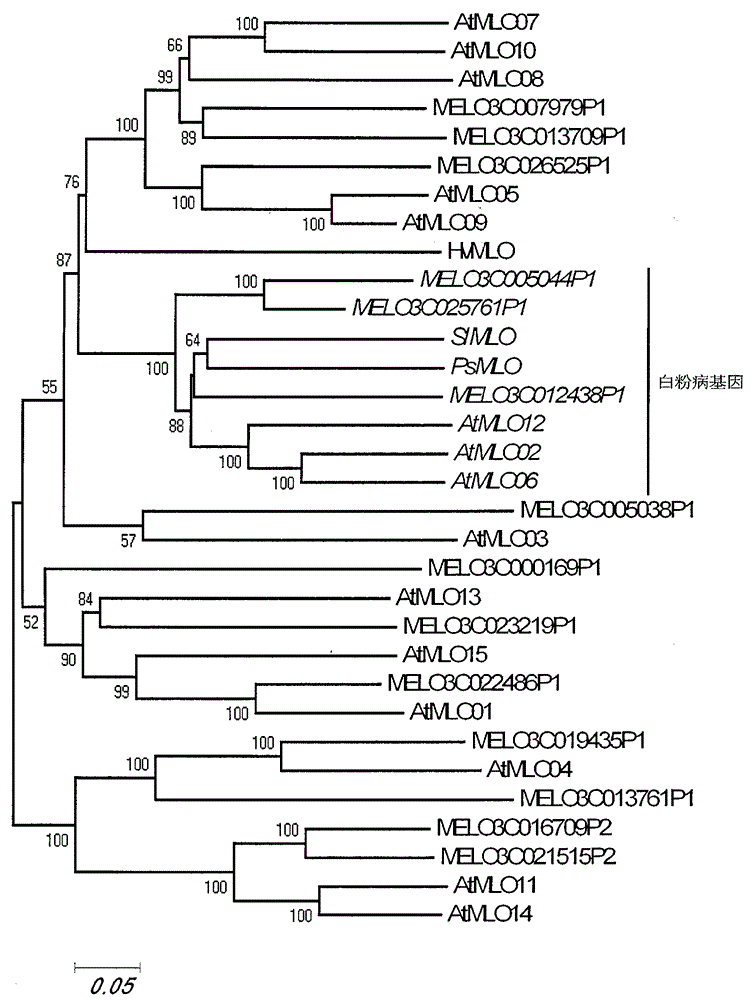
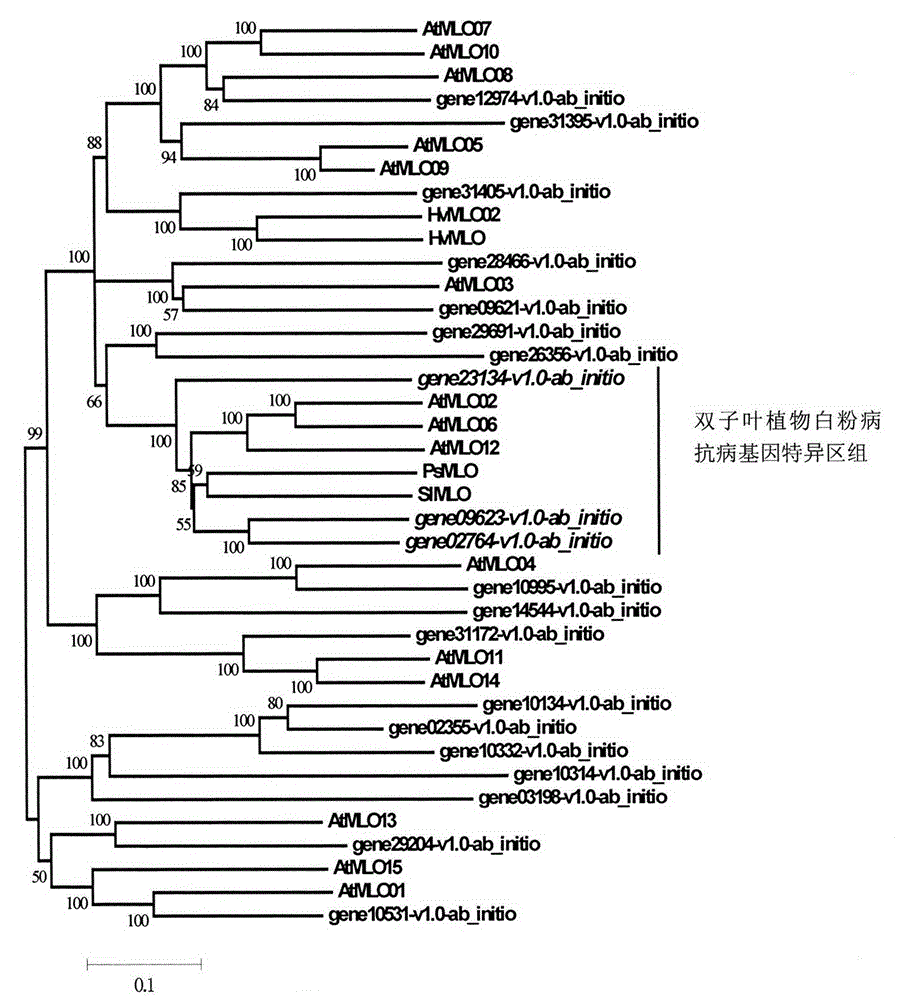
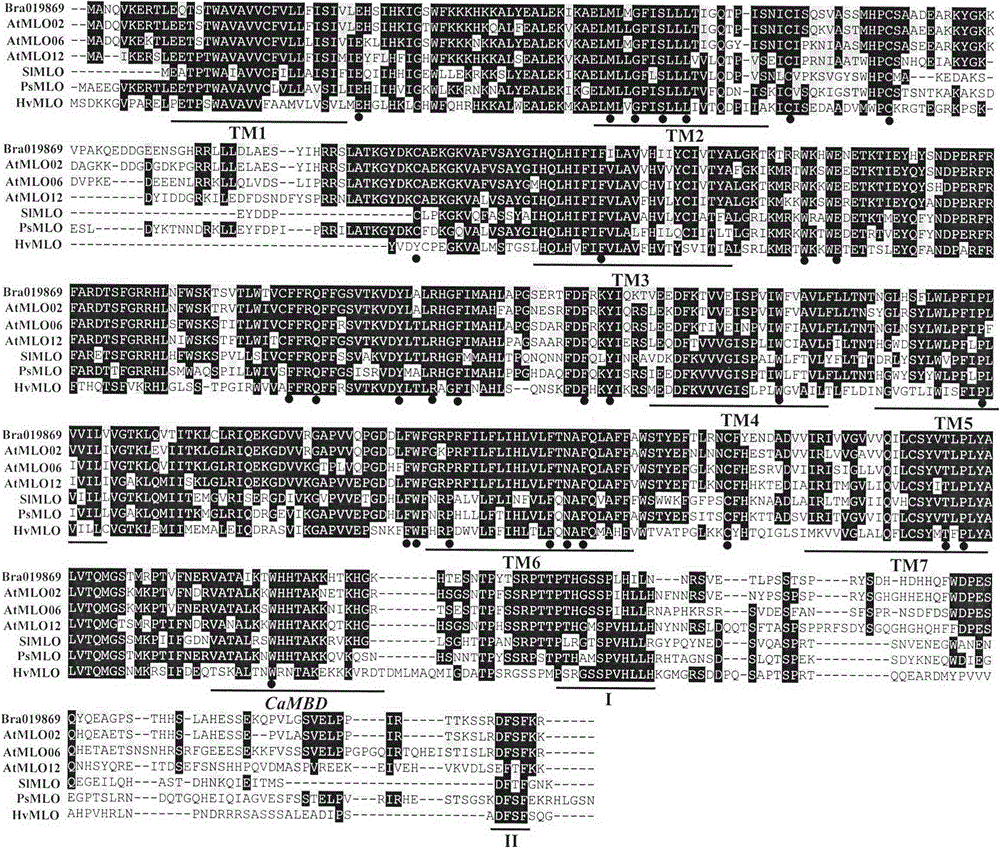
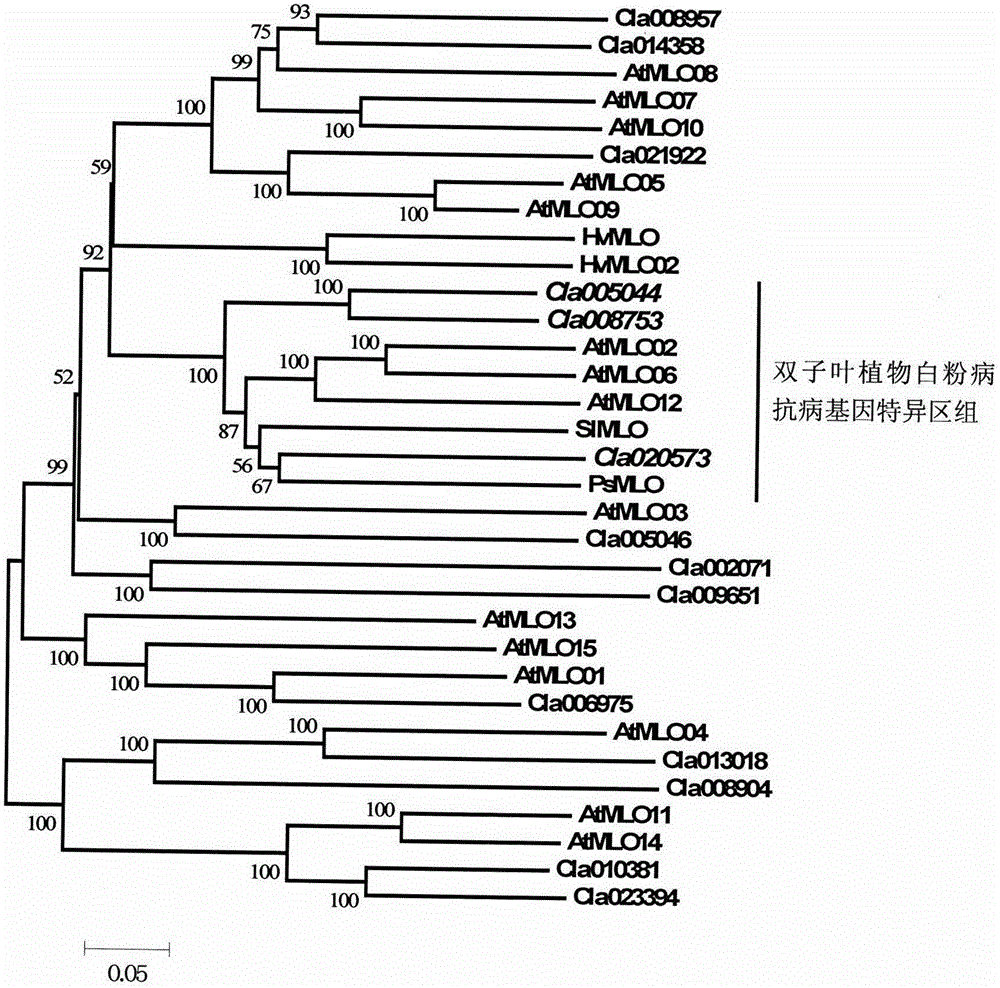
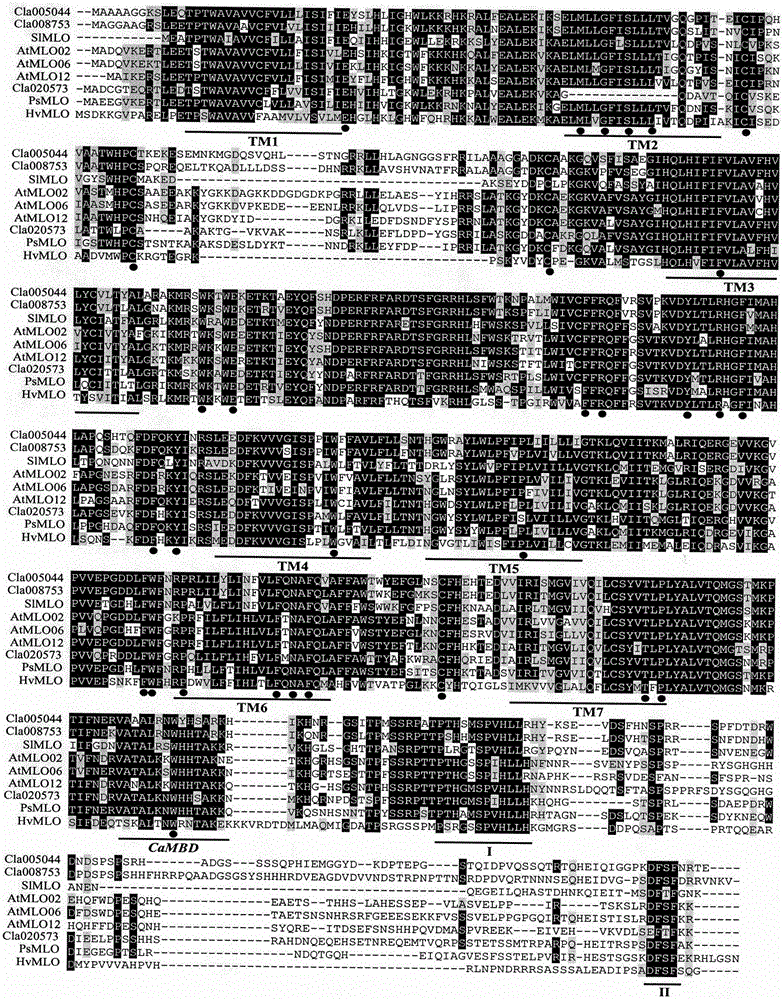
The invention relates to rapid identification of powdery mildew resistance Lotus corniculatus genes utilizing comparative genomics, relates to knowledge of subjects such as plant comparative genomics, genetics and bioinformatics, and belongs to the technical field of plant biotechnology. The rapid identification of powdery mildew resistance Lotus corniculatus genes utilizing the comparative genomics mainly includes the steps of firstly, downloading full genomic sequences of Lotus corniculatus and collecting MLO (mildew resistance locus o) genes; secondly, identifying the MLO genes; thirdly, identifying phylogenetic relationship of the MLO genes; and fourthly comparing the MLO powdery mildew resistance genes. By the rapid identification, mining cycle of the powdery mildew resistance genes is shortened, and the powdery mildew resistance genes can be identified quickly. Corresponding co-separation functional marks (SNP (single nucleotide polymorphism), Scar and the like) can be developed through candidate powdery mildew resistance genes identified, the rapid identification is also available for molecular marker-assisted selection of the powdery mildew resistance genes, and accuracy is high. The rapid identification can also be used with other molecular markers for resistance genes to create multiresistance breeding materials, and accordingly breeding period is shortened, breeding efficiency is improved, and basis for elaborating powdery mildew resistance Lotus corniculatus molecular mechanisms is laid.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
Application of bioinformatics in fast identifying powdery mildew resistance gene of carica papaya l
InactiveCN102719448AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesBiotechnologyPapaya family
The invention relates to fast identification of a powdery mildew resistance gene of carica papaya l, relating to subject knowledge including plant comparative genomics, genetics, bioinformatics and the like, belonging to the field of plant biotechnology sciences. Application of the invention mainly comprises the steps: 1) downloading complete genome sequence of the carica papaya l and acquiring MLO-type genes; 2) identifying the MLO-type genes; 3) researching phylogenetic relationships of the MLO-type genes; and 4) comparing MLO-type powdery mildew resistance genes. The invention has the advantages that discovery period of the powdery mildew resistance gene of the carica papaya l is effectively shortened, which assists in fast identification of the powdery mildew resistance gene; that identified candidate powdery mildew resistance genes are developed with corresponding co-segregation functional markers (SNP, SCAR, etc.), which can be used in fast auxiliary selection of molecular markers of the powdery mildew resistance gene with high accuracy; that other molecular markers of resistance genes can be combined to formulate multi-resistance breeding materials, which shortens breeding years and improves breeding yields; and that a foundation is laid to illustrate the powdery mildew resistance gene of the carica papaya l.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
Rapid identification of powdery mildew gene of Medicago truncatula by utilizing comparative genomics
InactiveCN102719445AQuick assist selectionImprove accuracyMicrobiological testing/measurementPlant peptidesBiotechnologyRapid identification
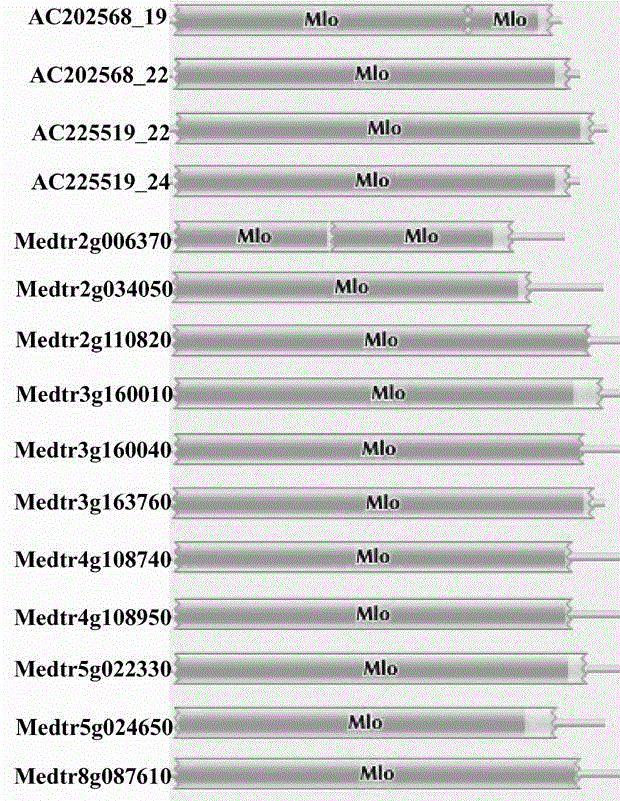
The present invention is rapid identification of a powdery mildew gene of Medicago truncatula, relates to discipline knowledge of plant comparative genomics, genetics and bioinformatics and belongs to the technical field of plant biotechnology. The invention mainly comprises the following steps: 1) download of Medicago truncatula genome sequence and collection of an MLO gene; 2) identification of MLO type gene; 3) MLO gene phylogenetic relationship; and 4) comparison of MLO type powdery mildew gene. The invention effectively shortens a mining cycle of the powdery mildew gene of Medicago truncatula and facilitates the rapid identification of the powdery mildew gene. The identified candidate powdery mildew genes can be used to develop corresponding coseparation functional markers (SNP and SCAR, etc.), and can also be used quickly for molecular marker assisted selection of powdery mildew resistant gene, and has high accuracy. Combined with other disease resistant gene molecular markers, the identified candidate powdery mildew genes can be used in development of multiresistance breeding material, so as to shorten the breeding period and improve breeding efficiency. The invention lays foundation to elaborate molecular mechanism of powdery mildew resistance of Medicago truncatula.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
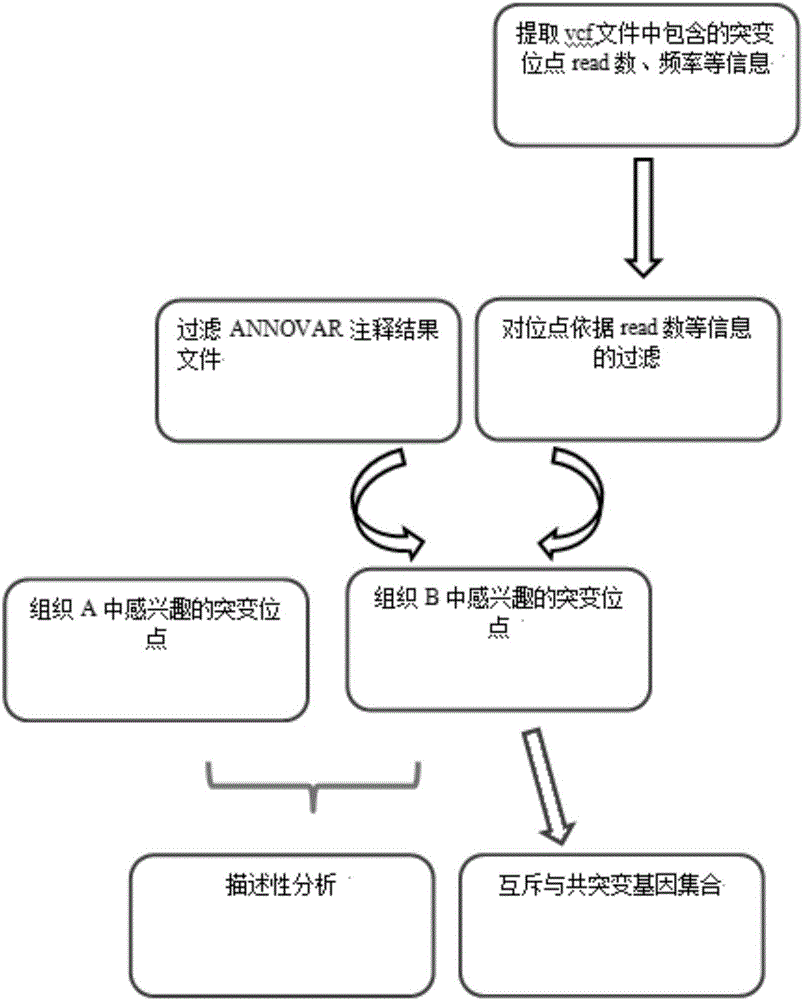
Tumor mutation site screening and mutual exclusion gene mining method
InactiveCN106021994AFacilitate biological interpretationHybridisationSpecial data processing applicationsWilms' tumorAnnotation
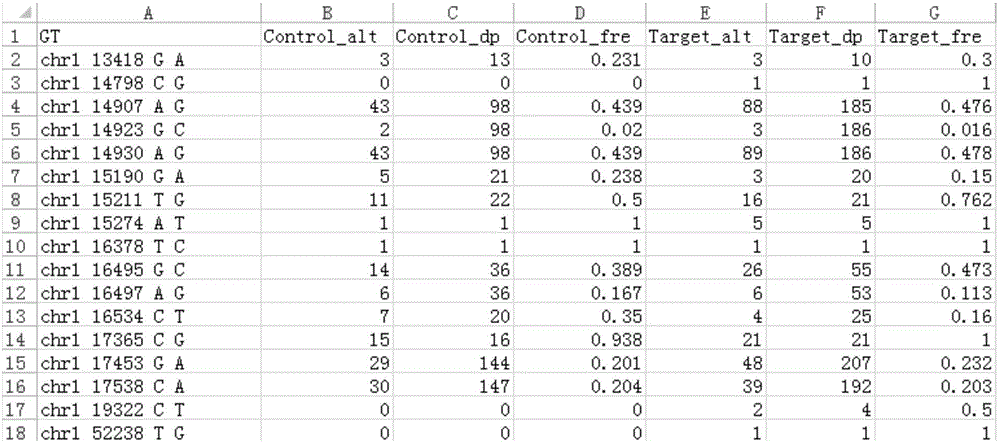
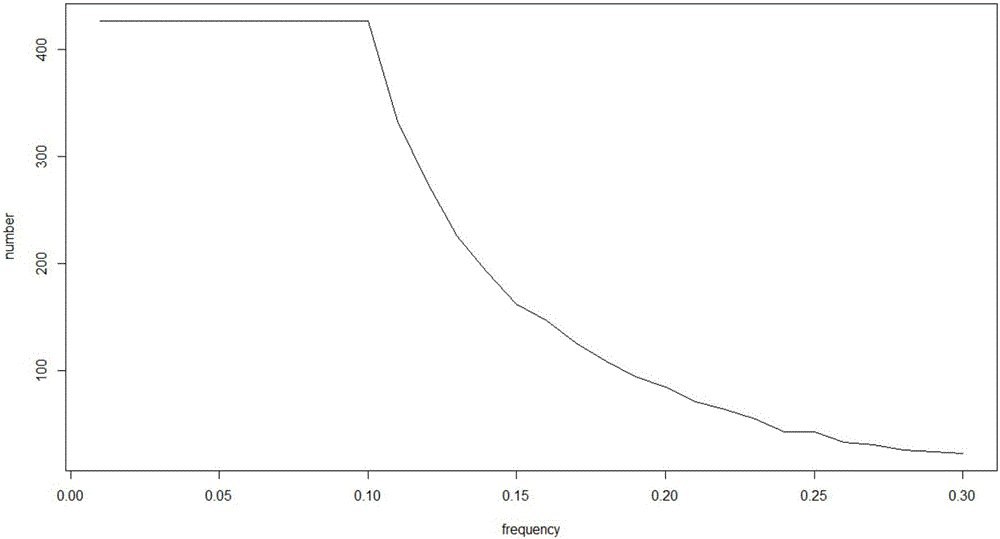
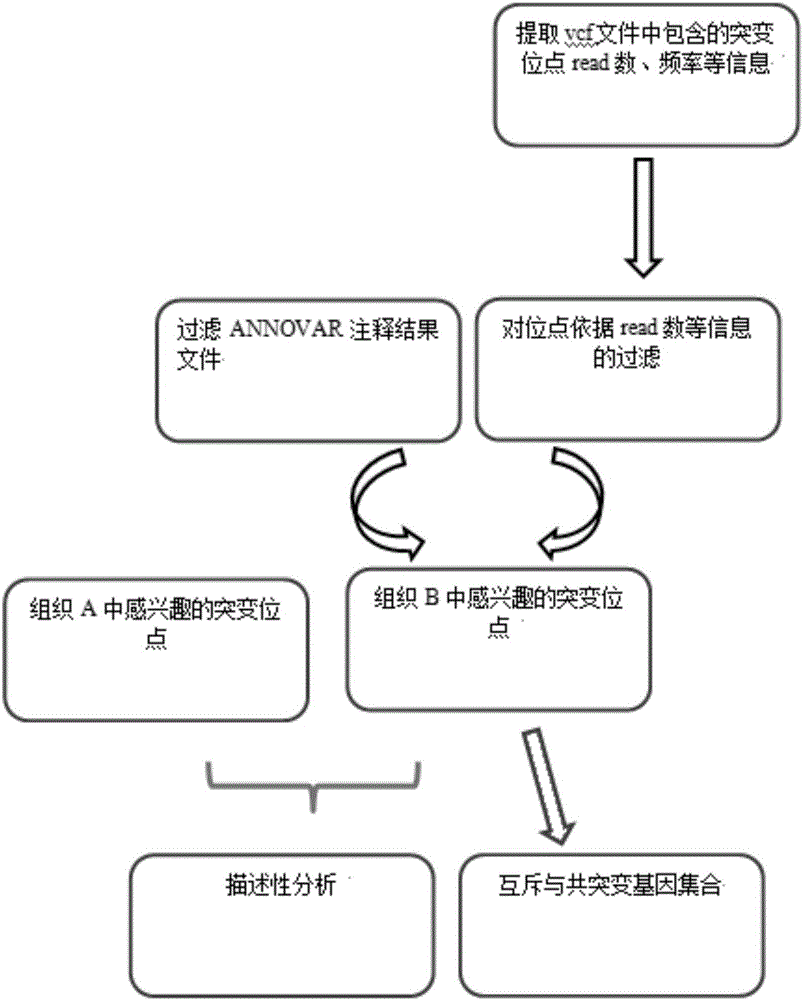
The invention provides a tumor mutation site screening and mutual exclusion gene mining method, which comprises the following steps: (1) filtering a vcf file and an output file of ANNOVAR annotation software; (2) carrying out the descriptive analysis of different experimental groups of mutation sites; (3) constructing a mutation gene matrix; and (4) carrying out mutual exclusion and joint mutation analysis on the generated mutation gene matrix on the basis of a Fisher exact test to determine a mutual exclusion and joint mutation gene. The mutation site is filtered by the annotation information of the mutation site and basic parameters including a sequencing read number, site sequencing depth and the like, and then, the obtained mutation site is subjected to the descriptive analysis of different experimental groups of mutation modes and the mining of a mutual exclusion and joint mutation gene set.
Owner:WANKANGYUAN TIANJIN GENE TECH CO LTD
Method for quickly identifying manihot esculenta mildew-resistance locus (MLO) gene by applying comparative genomics
InactiveCN102796745AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesDiseaseGenetics genomics
The invention discloses a method for quickly identifying a manihot esculenta mildew-resistance locus (MLO) gene, relates to knowledge of plant comparative genomics, genetics, bioinformatics and the like and belongs to the field of plant biotechnology science. The method mainly comprises the following steps of: 1) downloading a manihot esculenta whole genome sequence, and collecting the MLO gene; 2) identifying the MLO gene; 3) identifying the MLO gene according to the MLO gene phylogenetic relationship; and 4) comparing the MLO genes. By the method, the discovery cycle of the manihot esculenta MLO gene is shortened effectively, and the MLO gene can be quickly identified; corresponding coseparation functional markers (single nucleotide polymorphism (SNP) and specific combining ability (SCA)) can be developed by the identified candidate MLO gene, and the method can be quickly used for molecular marker auxiliary selection of the MLO gene, and is high in accuracy; by combining other disease-resistant gene molecular markers, multiresistance breeding materials can be prepared, the breeding cycle is shortened, and the breeding efficiency is improved; and the foundation is laid for elaborating a manihot esculenta MLO gene molecular mechanism.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
Calabash gourd anti-plague gene segment or gene marker and application
InactiveCN102703439AQuick assist selectionImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationNucleotideRapid identification
The invention mainly relates to a calabash gourd anti-plague gene segment or gene marker and application. Three calabash gourd anti-plague gene segments or gene markers are rapidly separated by adopting an anti-disease gene homologous sequence method, and by sequencing, the nucleotide lengths of the gene segments or gene markers are 510bp, 498bp and 501bp. According to the calabash gourd anti-plague gene segment or gene marker and application provided by the invention, the excavation period of the calabash gourd anti-plague gene is effectively shortened, and the rapid identification of plague gene is facilitated; the identified plague gene can be used for developing corresponding coseparation functional markers (SNP (single nucleotide polymorphism), SCAR (sequence-characterized amplified region) and the like), and can also be rapidly used for assisted selection of a molecule marker of the anti-plague gene, and the accuracy is high; development of multi-resistance breeding materials can be carried out by combining with other anti-disease gene molecule markers, the breeding time is shortened and the breeding efficiency is improved; and foundation is established for expounding the molecular mechanism of the anti-plague gene.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
Methods to design probes and primers
InactiveUS20090198479A1Enhances gene miningImprove efficiencyAnalogue computers for chemical processesSequence analysisGenomeBioinformatics
A web-based computational system and method for high-throughput gene mining and target identification and validation based on conserved functional blocks automates the generation of conserved regions of high sequence similarity using algorithms and optimizes probe design for fragment amplification from homologous genomes.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Tumor mutation site screening and mutual exclusion gene mining system
InactiveCN106022001AFacilitate biological interpretationBiostatisticsSpecial data processing applicationsGene miningAnnotation
The invention provides a tumor mutation site screening and mutual exclusion gene mining system. The system comprises a filtering module which is used for filtering a vcf file and an ANNOVAR software output file in an exome processing process; an analysis module which is used for carrying out description analysis on different experiment group mutation sites; a gathering module which is used for gathering mutation genes of each sample and establishing a mutation gene matrix according to an experiment group mutation gene list; and a mining module which is used for carrying out Fisher precise checking based mutual exclusion and co-mutation analysis on the generated mutation gene matrix, thereby determining mutual exclusion and co-mutation genes. According to system, the mutation sites are filtered according to basic parameters such as annotation information of the mutation sites, the sequencing read number and site sequencing depth; and description analysis of different experiment group mutation modes and mining of co-mutation and mutual exclusion gene sets are carried out on the obtained mutation sites.
Owner:WANKANGYUAN TIANJIN GENE TECH CO LTD
Rapid identification of anti-powdery-mildew gene of Brachypodium distachyon
InactiveCN102703462AShorten digging cycleRapid identificationMicrobiological testing/measurementPlant peptidesBiotechnologyBrachypodium sylvaticum
The invention relates to rapid identification of anti-powdery-mildew gene of Brachypodium distachyon, relates to the knowledge of plant comparative genomics, genetics, biological information and other subjects, and belongs to the scientific field of plant biotechnology. The rapid identification of anti-powdery-mildew gene of Brachypodium distachyon comprises the main steps of: (1) downloading full genome sequences of Brachypodium distachyon and collecting MLO (mildew resistance locus o) type gene; (2) identifying the MLO type gene; (3) analyzing the MLO type gene historical development relation; and (4) comparing the MLO type powdery mildew gene. According to the rapid identification of anti-powdery-mildew gene of Brachypodium distachyon provided by the invention, the excavation period of the anti-powdery-mildew gene of Brachypodium distachyon is effectively shortened, and the rapid identification of the powdery mildew gene is facilitated; the identified powdery mildew gene can be used for developing corresponding coseparation functional markers (SNP (single nucleotide polymorphism), SCAR (sequence-characterized amplified region), and the like), and can also be rapidly used for assisted selection of molecule markers of anti-powdery-mildew gene, and the accuracy is high; development of multi-resistance breeding materials can be carried out by combining with other anti-disease gene molecule markers, the breeding time is shortened and the breeding efficiency is improved; and foundation is established for expounding the molecular mechanism of the anti-powdery-mildew gene of Brachypodium distachyon.
Owner:常熟市支塘镇新盛技术咨询服务有限公司
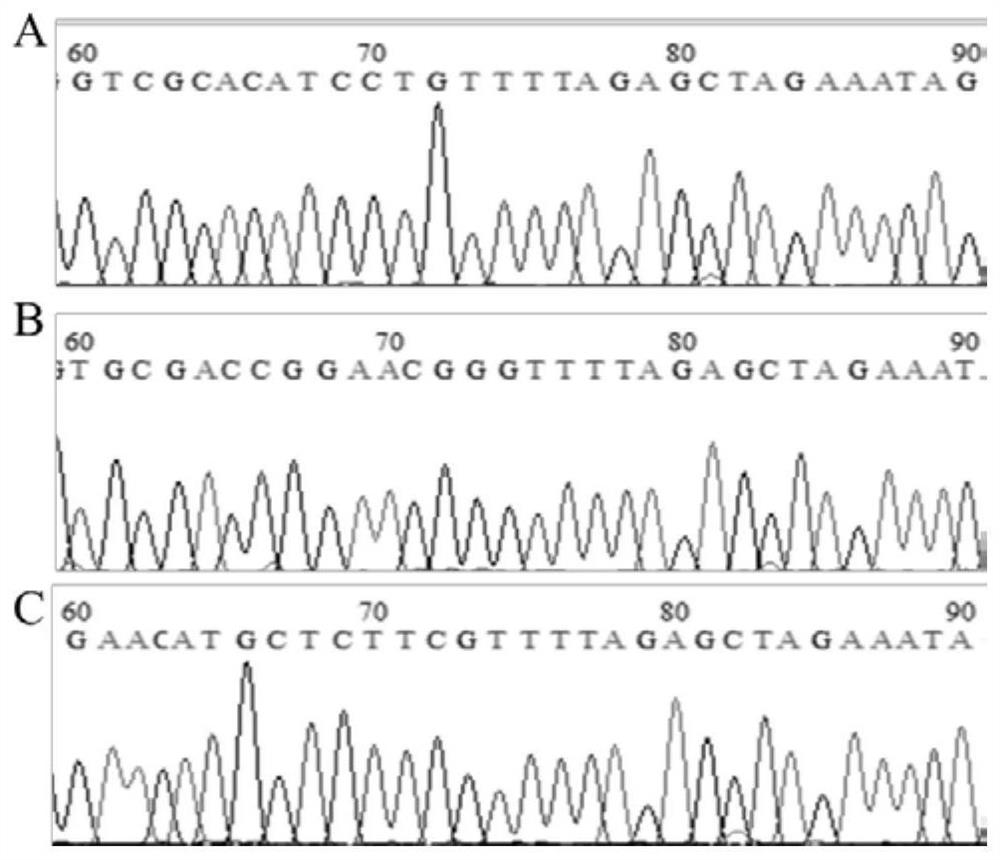
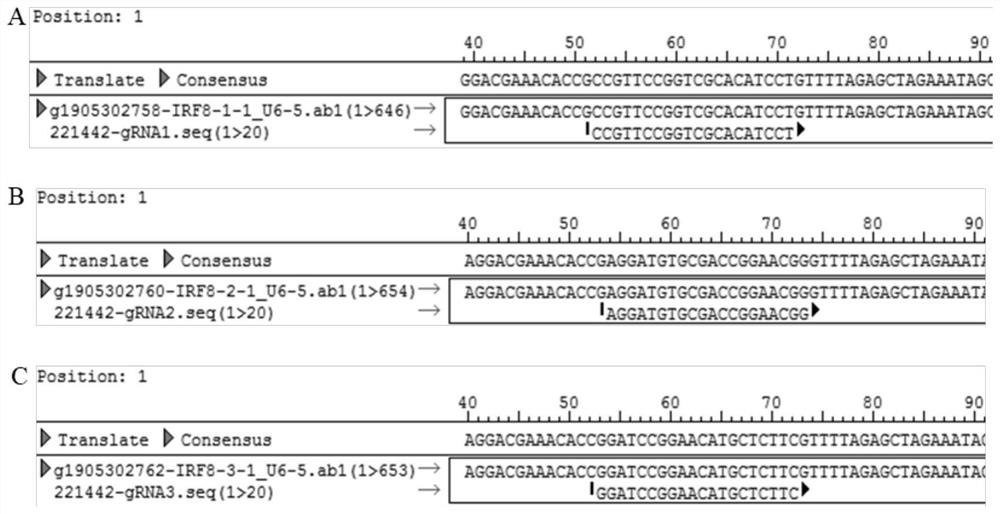
Cell line for knocking out porcine IRF8 gene based on CRISPR-Cas9 editing technology and construction method of cell line
PendingCN111607594AContribute to researchIdeal knockoutStable introduction of DNAPeptidesNegative strandDouble strand
The invention discloses a cell line for knocking out the porcine IRF8 gene based on a CRISPR-Cas9 editing technology and a construction method of the cell line. The cell line is prepared by adopting the following steps of: (1) design of a target site and synthesis of an sgRNA sequence: designing an sgRNA guide sequence according to the 5' end of a CDS region of the porcine IRF8 gene; (2) vector construction: annealing the plus-strand sgRNA sequence and the minus-strand sgRNA sequence to form double-stranded DNA; connecting the double-stranded DNA with a linearized pGK1.1 vector to obtain a positive targeting vector; and (3) cell transfection: carrying out mixed electrotransfection of the positive targeting vector into the target cell IPEC-J2 to obtain the IPEC-J2 cell with the IRF8 gene knocked out. The small intestine epithelial cell line with the IRF8 gene knocked out is established, and a more direct and effective research model can be provided for deep revealing of a diarrhea pathogen pathogenic mechanism, resistance gene mining and identification and cultivation and application of diarrhea-resistant transgenic pigs.
Owner:YANGZHOU UNIV
Gene mining system and method
InactiveUS20050196807A1Cell receptors/surface-antigens/surface-determinantsSugar derivativesBioinformaticsPhenotype
The present invention provides a system, method and apparatus for targeting gene sequences having one or more phenotypic characteristics using a computer. One or more phenotypic characteristics are selected. A gene sequence is then selected that is known to have the selected phenotypic characteristics. In addition, one or more databases containing cataloged gene sequences are selected. The selected gene sequence is compared to the cataloged gene sequences, and any cataloged gene sequences that contain a portion of the selected gene sequence are extracted. The selected gene sequence is aligned to each portion of the extracted gene sequence and the extracted gene sequences are prioritized based on the alignment of the selected gene sequence. At least one of the prioritized gene sequences is selected based on one or more phenotypic criteria. Finally, one or more degenerate primers are designed to target the selected-prioritized gene sequences.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Gene mining and application method of phospholipase D
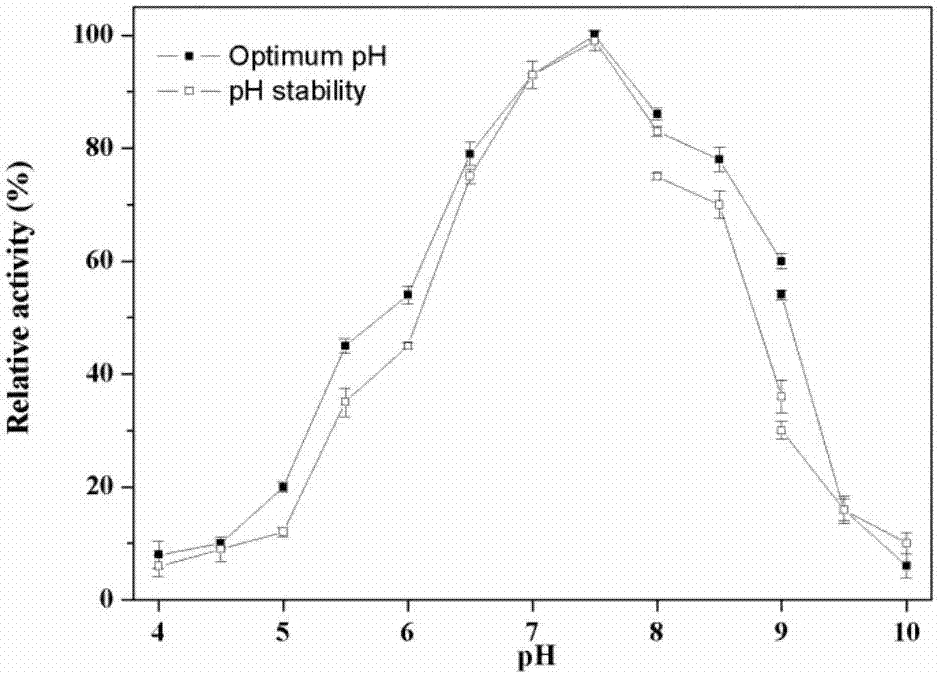
InactiveCN107164397AEfficient expressionShort fermentation cycleHydrolasesFermentationEscherichia coliReaction temperature
The invention provides a phospholipase D acquired based on a gene mining method, as well as a gene coding sequence and application method thereof. A phospholipase D gene has a full length of 1,614bp, encodes 538 amino acids, uses plasmid pET28a (+) as an expression vector, and uses E. coli BL21 (DE3) as an expression host, to realize efficient expression of phospholipase D. The phospholipase D gene is simple and convenient in gene mining method; a recombinant strain constructed by the mined target gene can be fermented and induced to excrete the phospholipase D. For the phospholipase D, an optimum reaction temperature is 60 DEG C; an optimum reaction pH is 7.5; and Ca<2+> has good activating and promoting effects on enzyme activity of the phospholipase D; the phospholipase D can modify phospholipids, and can catalyze substrates phosphatidylcholine and L-serine to synthesize phosphatidylserine, thereby having good application prospects in food, medicine and healthcare industries.
Owner:JIANGNAN UNIV
Application of L-threonine transaldolase to synthesis of florfenicol chiral intermediates
ActiveCN110540977AIncrease added valueSave separation and purificationTransferasesFermentationEscherichia coliBenzaldehyde
The invention provides application of L-threonine transaldolase to synthesis of florfenicol chiral intermediates. The L-threonine transaldolase is selected from any one of the following groups that (1) polypeptide contains an amino acid sequence shown in SEQ ID NO:1; (2) polypeptide contains an amino acid sequence greater than or equal to 90% homology shown in SEQ ID NO:1, and the polypeptide hascatalytic activity; and (3) derived polypeptide is formed by substitution, deletion or addition of 1-5 amino acid residues to the amino acid sequence shown in SEQ ID NO:1 and with the retention of catalytic activity. According to the application, new L-threonine transaldolase genes are screened through gene mining, the L-threonine transaldolase derived from recombinant escherichia coli is expressed by adopting a genetic engineering method, methylsulfonyl benzaldehyde and the L-threonine transaldolase are used as raw materials, (2S,3R)-methylsulfonyl phenylserine is synthesized through a wholecell catalyzed reaction, fluorfenicol key chiral synthesis blocks can be obtained by a one-step reaction under the room temperature and pressure, environmental protection is achieved, and the application is an environment-friendly biosynthetic pathway.
Owner:福建昌生生物科技发展有限公司
Rapid Identification of pear powdery mildew resistance gene
InactiveCN104561027AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementGenetic engineeringAgricultural scienceRapid identification
The invention relates to rapid identification of a pear powdery mildew resistance gene, and relates to the subject knowledge of plant comparative genomics, genetics and bioinformatics, belonging to the field of science of biotechnology. The rapid identification of the pear powdery mildew resistance gene comprises the following main steps of: 1) downloading of a complete genome sequence of a pear and collection of an MLO gene; 2) identification of the MLO gene; 3) MLO gene phylogenetic relationship; and 4) comparison of an MLO powdery mildew gene. The rapid identification of the pear powdery mildew resistance gene, provided by the invention, has the advantages of effectively shortening the pear powdery mildew gene mining period, and being favorable for the rapid identification of the powdery mildew gene; corresponding co-segregation functional markers (SNP, SCAR and the like) can be developed through the identification of candidate powdery mildew genes, and the co-segregation functional markers can be rapidly used for the molecular marker-assisted selection of the powdery mildew resistance gene, so that the accuracy is high; by combining other powdery mildew resistance gene molecular markers, a multi-resistance breeding material can be created, the breeding period can be shortened, and the breeding efficiency can be improved; a foundation is laid for explaining a pear powdery mildew resistance molecular mechanism.
Owner:CHANGSHU CITY DONGBANG DONGDUN VEGETABLE PROFESSION COOP
Rapid identification of soybean anti-powdery mildew gene by using candidate gene strategy
InactiveCN104593481AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementBiostatisticsCandidate Gene Association StudyRapid identification
The invention provides rapid identification of soybean anti-powdery mildew gene, relates to knowledge in the disciplines of plant comparative genomics, genetics and bioinformatics, and belongs to the scientific field of plant biotechnology. The invention includes the steps of: 1) downloading the soybean whole genome sequence and acquiring MLO type gene; 2) identifying the MLO genotype; 3) identifying MLO gene phylogenetic relationship; and 4) comparing MLO-type powdery mildew gene. The invention effectively shortens the gene mining cycle for soybean powdery mildew, and is conducive to the rapid identification of anti-powdery mildew gene; corresponding coseparation functional markers (SNP, scar, etc.) are developed through the identified candidate powdery mildew genes, and the identified candidate powdery mildew genes can also be used for fast molecular marker assisted selection of anti-powdery mildew genes and have high accuracy; and combined with other resistant gene molecular markers, multi-resistance breeding material can be created, so as to shorten the breeding period and improve breeding efficiency. Therefore, the invention lays foundation for the explanation of powdery mildew resistant molecular mechanism of soybean.
Owner:JIANGSU CHANGSHU MODERN AGRI IND PARK DEV
Multi-omics-based mining method for key gene of fat deposition trait of Nanyang black pigs
The invention discloses a multi-omics-based mining method for a key gene of a fat deposition trait of Nanyang black pigs. According to the method, in order to solve the problem of the existing methodsthat the accuracy of mining and identifying of swine fat deposition trait genes is low, transcriptomes of musculi longissimus dorsi of six pairs of full-sib or half-sib Nanyang black pig individualsof 6 months old with extreme back fat thickness and intramuscular fat content are sequenced by using a high-flux sequencing technology, important fat deposition candidate genes are sought and identified on the basis of phenotype determination and under the assistance of bioinformatics analysis, meanwhile, two groups of Nanyang black pigs are subjected to genome resequencing to find out important fat deposition candidate difference genes, between-group remarkable-difference-expression genes are screened through integrating transcriptome data, re-sequenced data and QTL database information and using bioinformatics and functional genomics methods, and gene functions and signal channels of the between-group remarkable-difference-expression genes are predicted.
Owner:NANYANG NORMAL UNIV
Gene mining method combining functional sequence and structural simulation, NADH preference type glufosinate-ammonium dehydrogenase mutant and application
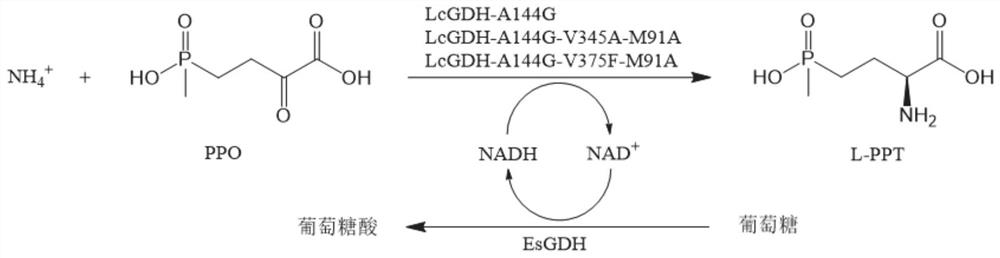
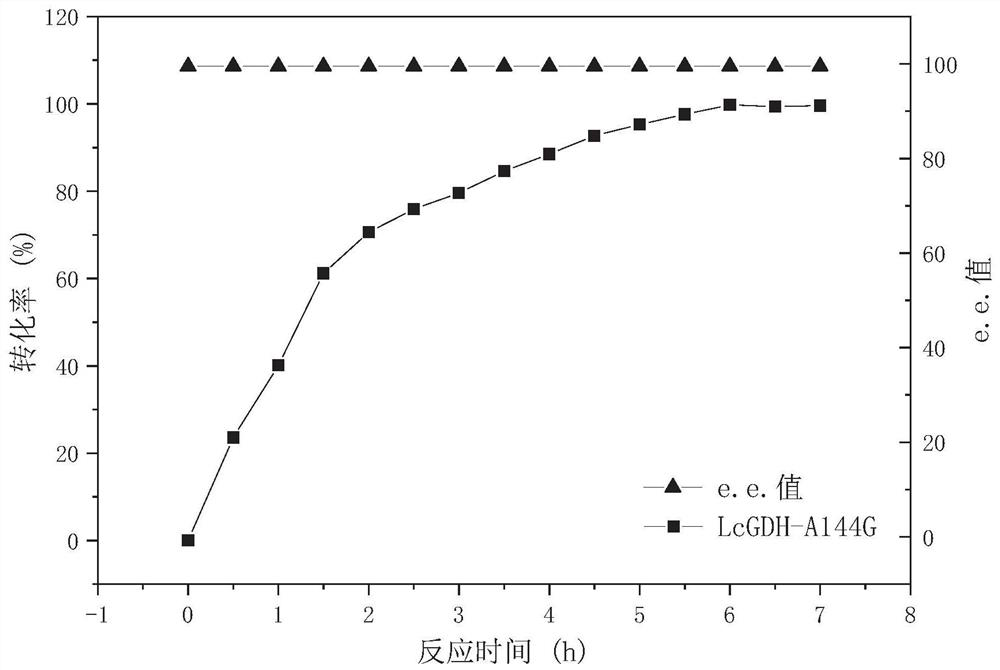
PendingCN112908417AHigh catalytic efficiencyImprove conversion rateProteomicsGenomicsEnzyme structureProtein structure
The invention discloses a gene mining method combining a functional sequence and structural simulation, an NADH preference type glufosinate-ammonium dehydrogenase mutant and an application. The gene mining method comprises the following steps: (1) analyzing a characteristic sequence of NADH type glutamate dehydrogenase; (2) searching a gene pool according to the characteristic sequence; (3) performing clustering analysis and protein structure simulation on the searched genes; and (4) selecting a gene with high gene polymerization degree and a protein structure similar to that of known glufosinate-ammonium dehydrogenase as a candidate gene. The method comprises the following steps: carrying out gene mining to obtain wild glufosinate-ammonium dehydrogenase which is derived from Lysinibacillus composti and has an amino acid sequence shown as SEQ ID No.2, carrying out mutation screening to obtain an NADH preference type glufosinate-ammonium dehydrogenase mutant, and selecting a mutation site from one of the following: (1) A144G-V375F-M91A; (2) A144G-V345A-M91A; and (3) A144G. The mutant enzyme can be subjected to catalytic reaction by using cheap coenzyme NAD.
Owner:ZHEJIANG UNIV OF TECH
Bottle gourd virus resistance-related gene segment or gene markers and application of gene segments or gene markers
InactiveCN104561029AQuick assist selectionImprove accuracyMicrobiological testing/measurementFermentationRapid identificationRelated gene
The invention mainly relates to bottle gourd virus resistance-related gene segments or gene markers and application thereof. A disease-resistant gene homologous sequence method is mainly adopted for rapidly separating three bottle gourd virus resistance-related gene segments or gene markers, sequencing is carried out, and lengths of nucleotides of the gene segment or gene markers are 510bp, 510bp and 504bp respectively. The bottle gourd virus resistance-related gene segments or gene markers have the advantages that a bottle gourd virus resistance-related gene mining period is effectively shortened, and rapid identification of a virus disease gene is facilitated; corresponding coseparation functional markers (SNP, SCAR and the like) are developed by virtue of the identified virus disease gene, the identified virus disease gene also can be used for virus disease resistant gene molecular marker-assisted selection rapidly with high accuracy; by combining with other disease-resistant gene molecular markers, a multi-resistance breeding material can be created, breeding cycle can be shortened, and breeding efficiency can be improved; and a foundation is laid for explaining a bottle gourd virus resistance-related gene molecular mechanism.
Owner:CHANGSHU ZHILIN FRUITS & VEGETABLES PROFESSIONAL COOP
Rapid identification of MLO (Mycoplasma Like Organism) powdery mildew resistant genes of melon
InactiveCN104561024AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementFermentationRapid identificationOrganism
The invention relates to rapid identification of MLO (Mycoplasma Like Organism) powdery mildew resistant genes of a melon, relates to comparative genomics of plants, genetics, bioinformatics and other disciplinary knowledge, and belongs to the field of biotechnology science of plants. The rapid identification is mainly carried by the following steps: 1) downloading the whole genome sequence of the melon, and acquiring MLO genes; 2) identifying the MLO genes; 3) obtaining the phylogenetic relationship of MLO genes; 4) comparing MLO powdery mildew resistant genes. By virtue of adopting the rapid identification, the powdery mildew resistant genes mining cycle of the melon can be effectively reduced, which is beneficial to the rapid identification of powdery mildew resistant genes; the identified candidate powdery mildew resistant genes are utilized to develop corresponding coseparation functional markers (such as SNR and SCAR) and can also be quickly used for assistant selection of molecular markers resistant to powdery mildew resistant genes, and the accuracy is high; in combination with other anti-disease gene molecular markers, a plurality of resistive breeding materials can be created, the breeding periods can be reduced, and the breeding efficiency can be increased; the foundation is provided for describing a powdery mildew resistant molecular mechanisms of the melon.
Owner:南农大(常熟)新农村发展研究院有限公司
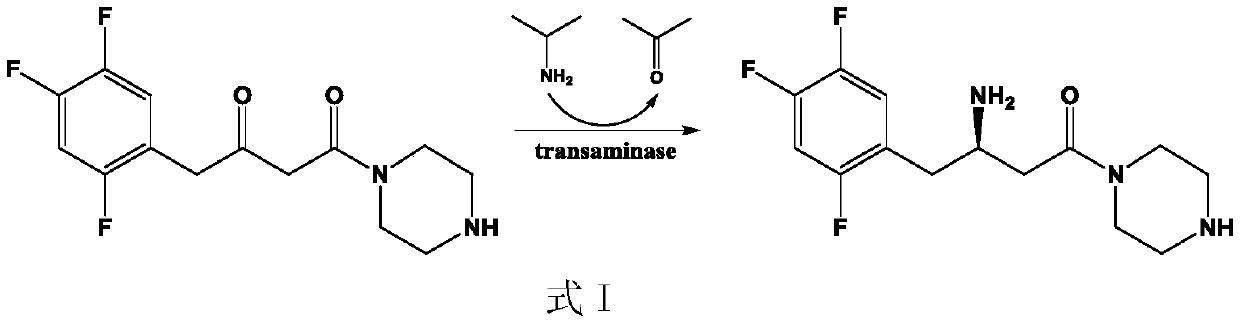
(R)-omega-transaminase mutant and application thereof
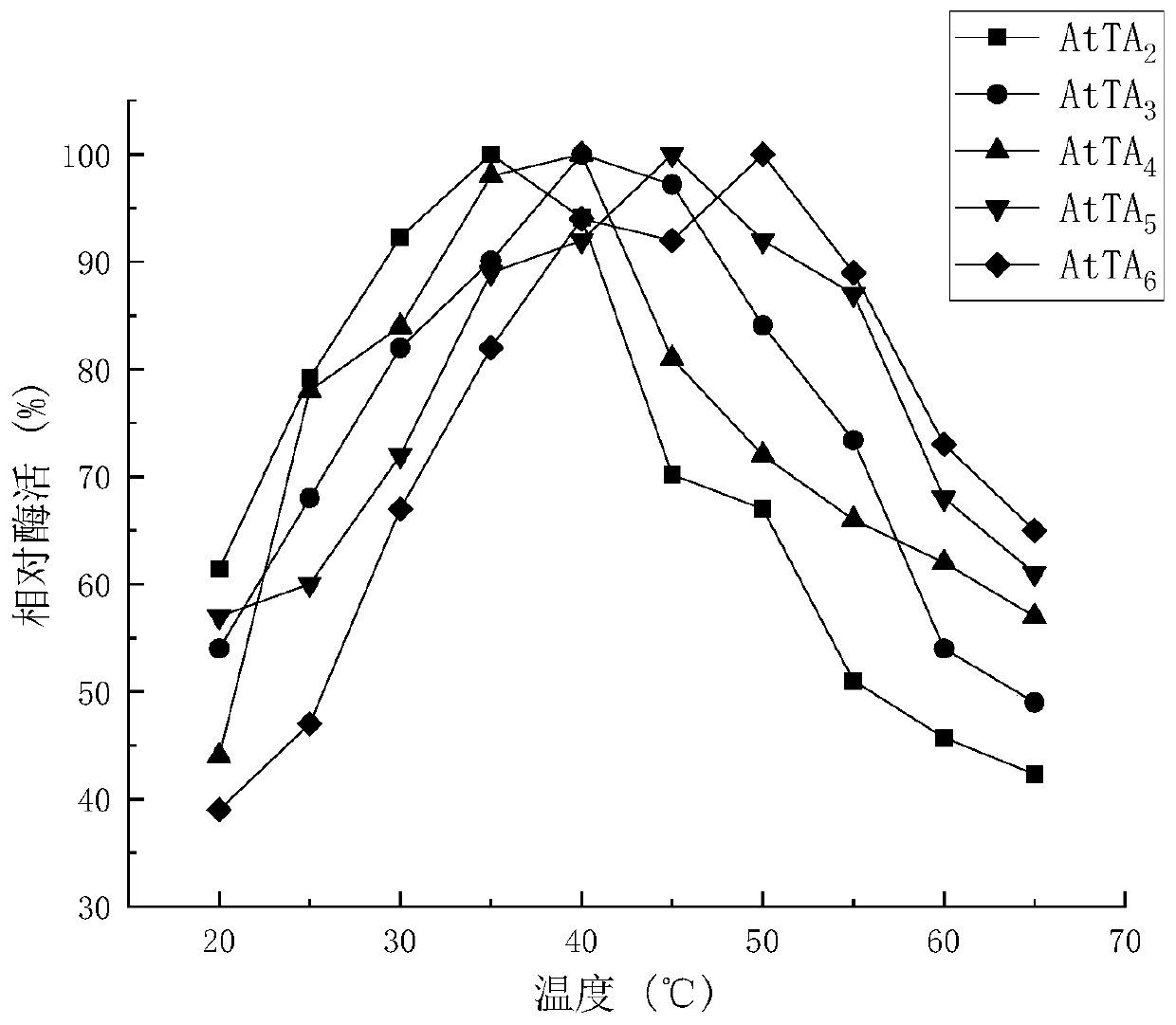
ActiveCN111411094AIncrease enzyme activityIncreased substrate toleranceBacteriaTransferasesArginineAcyl group
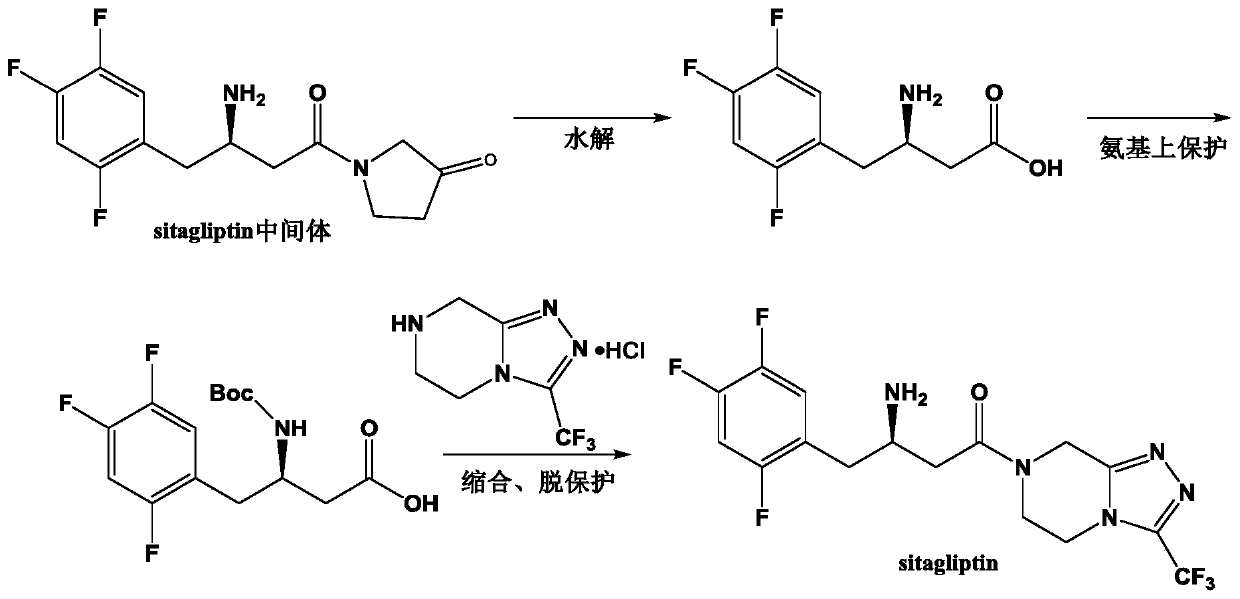
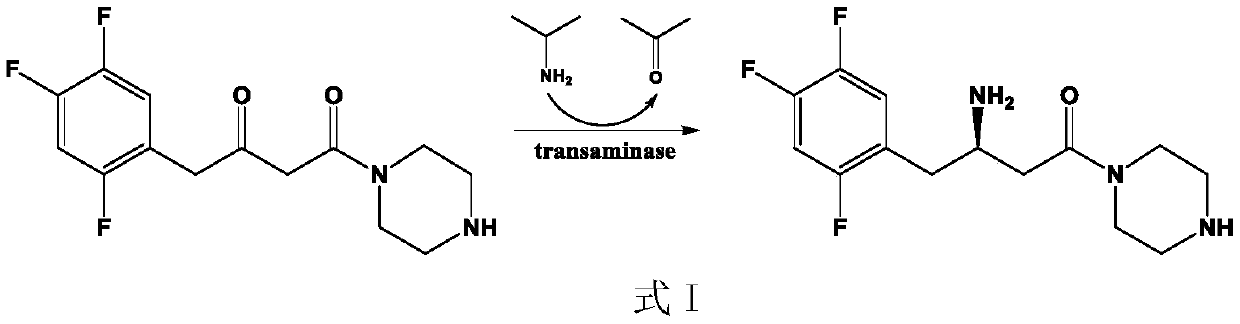
The invention discloses a (R)-omega-transaminase mutant and application thereof. The mutant is obtained by multi-point mutation of leucine at 182, arginine at 79, glutamine at 51, valine at 149, leucine at 235 and glycine at 216 in an amino acid sequence shown by SEQ ID NO.1. New (R)-omega-TA enzyme is screened through a gene mining technology in the invention; furthermore, molecular modificationis carried out through a protein engineering technology; a (R)-omega-TA mutant catalysis having high enzyme activity, high substrate tolerance and high stereoselectivity is obtained; the mutant can biologically catalyze precursor ketone analogue 1-(3-oxygen pyrrolidine-1-yl)-4-(2,4,5-trifluorophenyl)-1,3-butanedione synthesis sitagliptin intermediate (R)-1-[3-amino-4-(2,4,5-trifluorophenyl) butyryl] pyrrole-3-ketone; furthermore, the conversion rate is relatively high; the conversion rate can be up to 95.4%; and thus, the (R)-omega-transaminase mutant and application thereof in the invention have milestone meaning for breaking through a sitagliptin biocatalysis preparation technology.
Owner:ZHEJIANG UNIV OF TECH +2
Method for constructing metagenomic Fosmid library of soil microorganisms in tropical rainforest
InactiveCN107475244AEasy to digEfficient use ofBacteriaMicroorganism based processesFosmidLow voltage
The invention belongs to the field of microorganisms, and discloses a method for constructing a metagenomic Fosmid library of soil microorganisms in a tropical rainforest. The method comprises the following steps: washing by using cross-linked polyvinyl pyrrolidone (PVPP), removing impurities, and then adopting a lysozyme-protease K-SDS-CTAB lysis method to extract and purify deoxyribonucleic acid (DNA) in large fragments of soil microbial genomes; carrying out agarose gel low-voltage electrophoresis detection and pulsed electrophoresis detection; carrying out tail end repairing on the metagenomic DNA, cutting off a marker and a small sample, and using ligase to enable a recycled product to be ligated with pCC1FOS of a Fosmid carrier; carrying out a ligation reaction within 2h; packing a ligation product with a lambda packaging extract, carrying out transfection by using an EPI300-T1R strain, coating an LB flat plate, which contains 12.5mu g / ml of chloramphenicol, with a product of the transfection, and culturing overnight at the temperature of 37 DEG C. The method provided by the invention reflects that in the extreme environment of the tropical rainforest, the microorganisms and genetic resources thereof are extremely rich and abundant; the method is beneficial to the excavation and utilization of functional genes of the microorganisms.
Owner:INST OF PLANT PROTECTION HAINAN ACADEMY OF AGRI SCI
Gene mining system and method
InactiveUS7349811B2Cell receptors/surface-antigens/surface-determinantsSugar derivativesBioinformaticsPhenotype
The present invention provides a system, method and apparatus for targeting gene sequences having one or more phenotypic characteristics using a computer. One or more phenotypic characteristics are selected. A gene sequence is then selected that is known to have the selected phenotypic characteristics. In addition, one or more databases containing cataloged gene sequences are selected. The selected gene sequence is compared to the cataloged gene sequences, and any cataloged gene sequences that contain a portion of the selected gene sequence are extracted. The selected gene sequence is aligned to each portion of the extracted gene sequence and the extracted gene sequences are prioritized based on the alignment of the selected gene sequence. At least one of the prioritized gene sequences is selected based on one or more phenotypic criteria. Finally, one or more degenerate primers are designed to target the selected-prioritized gene sequences.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Rapid identification of strawberry powdery mildew gene by using comparative genomics
InactiveCN104611407AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementPlant peptidesFragariaAgricultural science
Owner:CHANGSHU CITY BEIBANG TOWN BEIGANG VEGETABLE SPECIALIZED COOP
Application of comparative genomics to rapid identification of phaseolus vulgaris mildew resistance locus o gene
InactiveCN104593480AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementBiostatisticsRapid identificationOrganism
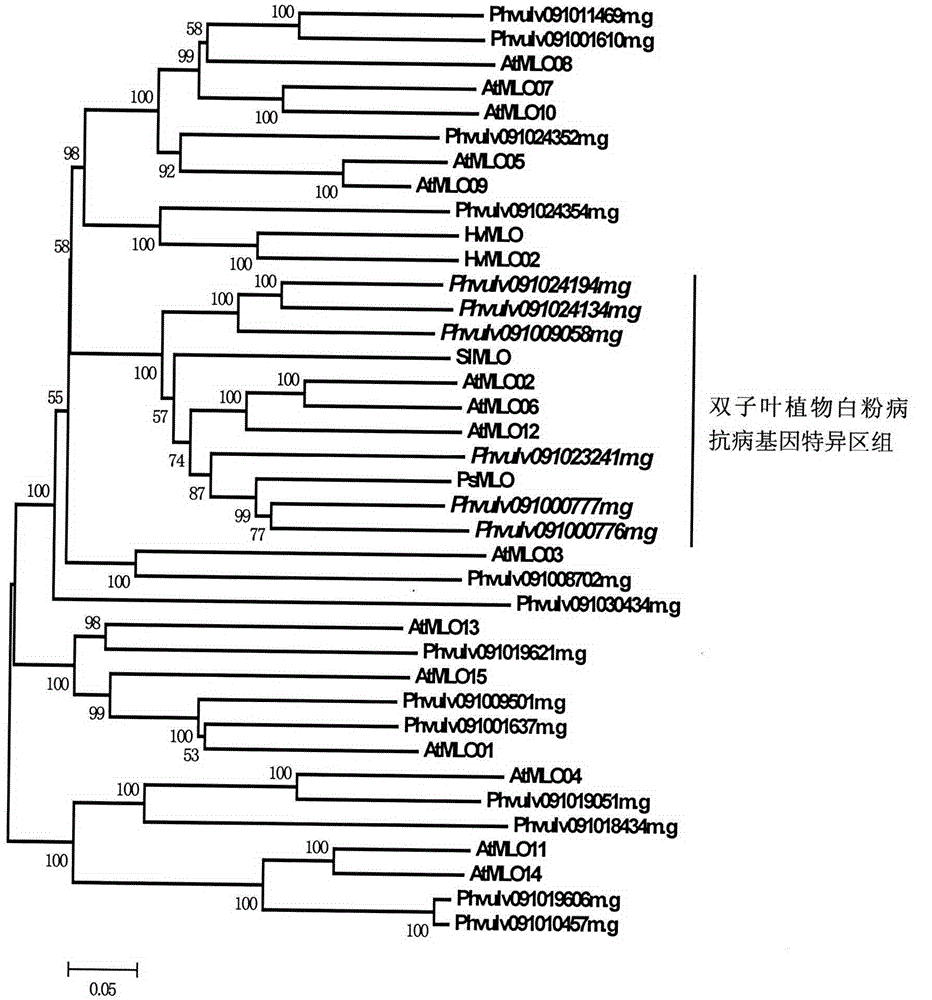
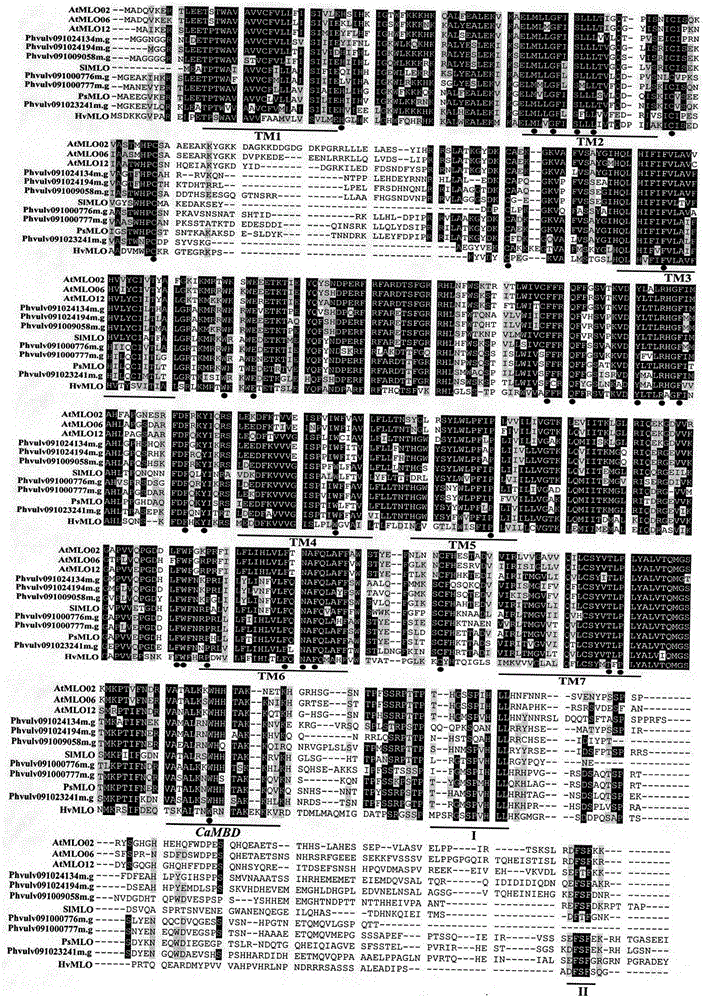
Relating to plant comparative genomics, genetics, bioinformatics and other disciplinary knowledge, the invention belongs to the scientific field of plant biotechnology, and discloses rapid identification of phaseolus vulgaris mildew resistance locus o gene. The main steps include: 1) download of phaseolus vulgaris whole genome sequence and acquisition of MLO type gene; 2) identification of MLO type gene; 3) MLO type gene phylogenetic relationship; and 4) comparison of MLO type powdery mildew. The invention effectively shortens the mining cycle of phaseolus vulgaris powdery mildew gene, and is conducive to rapid identification of powdery mildew gene. Corresponding coseparation functional markers (SNP, SCAR and the like) are developed through identified candidate powdery mildew genes, which also can be used for molecular marker-assisted selection of mildew resistance locus o gene and have high accuracy. Multi-resistance breeding materials can be created by combining other resistant gene molecular markers, the breeding period can be shortened, and the breeding efficiency is improved, thus laying the foundation for elaborating the phaseolus vulgaris mildew resistance molecular mechanism.
Owner:JIANGSU CHANGSHU MODERN AGRI IND PARK DEV
Rapid identification of powdery mildew resistant gene of Chinese cabbage by using comparative genomics
InactiveCN104593379AShorten digging cycleRapid identificationMicrobiological testing/measurementFermentationRapid identificationGenotype
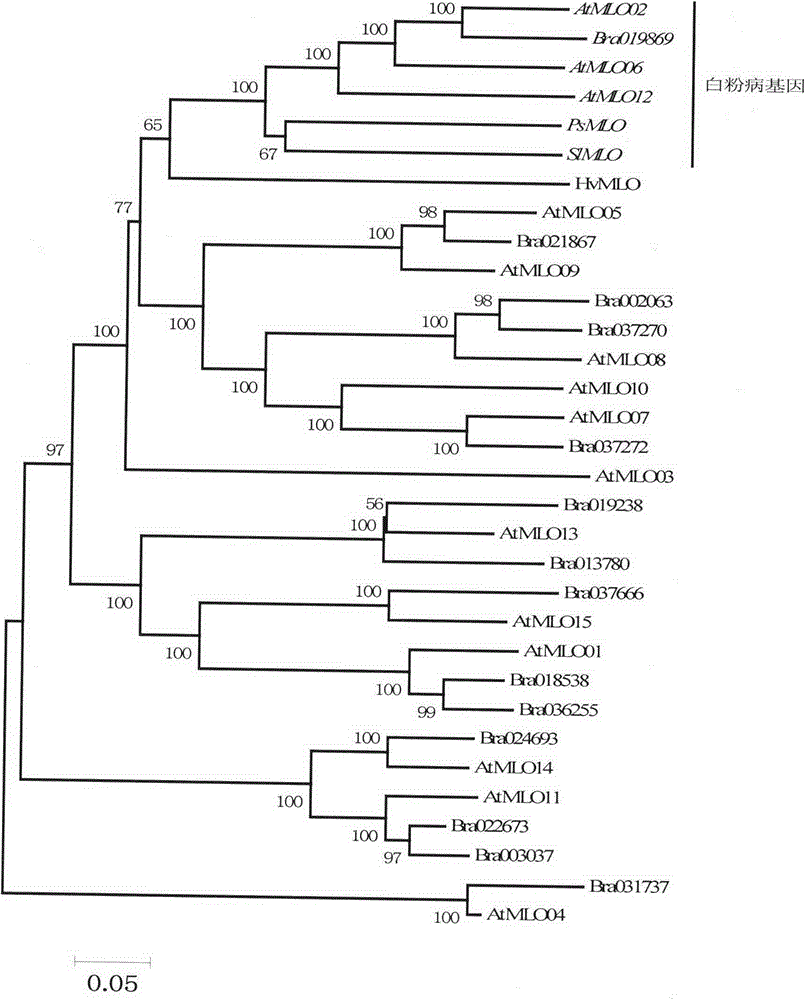
The invention provides rapid identification of powdery mildew resistant gene of Chinese cabbage, relates to knowledge in the disciplines of plant comparative genomics, genetics and bioinformatics, and belongs to the scientific field of plant biotechnology. The invention includes the steps of: 1) downloading the whole genome sequence of Chinese cabbage and acquiring MLO type gene; 2) identifying the MLO genotype; 3) identifying MLO gene phylogenetic relationship; and 4) comparing MLO-type powdery mildew gene. The invention effectively shortens the gene mining cycle for Chinese cabbage powdery mildew, and is conducive to the rapid identification of powdery mildew resistant gene; corresponding coseparation functional markers (SNP, SCAR, etc.) are developed through the identified candidate powdery mildew genes, and the identified candidate powdery mildew genes can also be used for fast molecular marker assisted selection of powdery mildew resistant genes and have high accuracy; combined with other resistant gene molecular markers, multi-resistance breeding material can be created, so as to shorten the breeding period and improve breeding efficiency. Therefore, the invention lays foundation for the explanation of powdery mildew resistant molecular mechanism of Chinese cabbage.
Owner:JIANGSU CHANGSHU MODERN AGRI IND PARK DEV
Rapid identification of watermelon MLO type anti-powdery mildew gene
InactiveCN104611408AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementPlant peptidesCo segregationMarker-assisted selection
The present invention discloses rapid identification of watermelon anti-powdery mildew gene, relates to knowledge of plant comparative genomics, genetics, bioinformatics and other disciplines, and belongs to the plant biotechnology science field. The main steps of the invention comprise: 1) downloading the complete genome sequence of the watermelon, and collecting MLO type gene; 2) identifying the MLO type gene; 3) achieving the MLO type gene phylogeny relationship; and 4) comparing the MLO type powdery mildew gene. According to the invention, the watermelon powdery mildew gene finding period is effectively shortened so as to easily achieve rapid powdery mildew gene identification; the corresponding co-segregation function marker (SNP, SCAR and the like) is developed through the identified candidate powdery mildew gene, the watermelon anti-powdery mildew gene can further be rapidly used for the molecule marker-assisted selection of the anti-powdery mildew gene, and the accuracy is high; the watermelon anti-powdery mildew gene can be combined with other disease resistance gene molecule markers so as to create multi-resistance breeding materials, shorten the breeding period, and improve the breeding efficiency; and the foundation is established for exposition of the watermelon anti-powdery mildew molecule mechanism.
Owner:CHANGSHU CITY BEIBANG TOWN BEIGANG VEGETABLE SPECIALIZED COOP
Quick identification of powdery mildew resistant gene for barley
InactiveCN104561032AShorten digging cycleImprove identification efficiencyMicrobiological testing/measurementGenetic engineeringResistant genesOrganism
The invention provides quick identification of a powdery mildew resistant gene for barley, relates to subject knowledge of plant comparative genomics, genetics, bioinformatics and the like and belongs to the field of plant biotechnology science. The quick identification mainly comprises the following steps: (1) downloading a barley whole-genome sequence and acquiring an MLO gene; (2) identifying the MLO gene; (3) establishing a phylogenetic relationship of the MLO gene; and (4) comparing an MLO powdery mildew gene. According to the quick identification, the mining period of the barley powdery mildew gene is effectively shortened to facilitate quick identification of the powdery mildew gene; the corresponding coseparation functional markers (SNP, SCAR and the like) are developed through the identified candidate powdery mildew gene and quickly used for molecular marker auxiliary selection of the powdery mildew resistant gene, and the accuracy is high; through combination with other anti-disease gene molecular markers, creation of a multi-resistant breeding material can be carried out, the breeding period is shortened, and the breeding efficiency is improved; and a foundation is laid for stating the barley powdery mildew resistance molecular mechanism.
Owner:CHANGSHU CITY DONGBANG TOWN LIMU VEGETABLE PROFESSION COOP
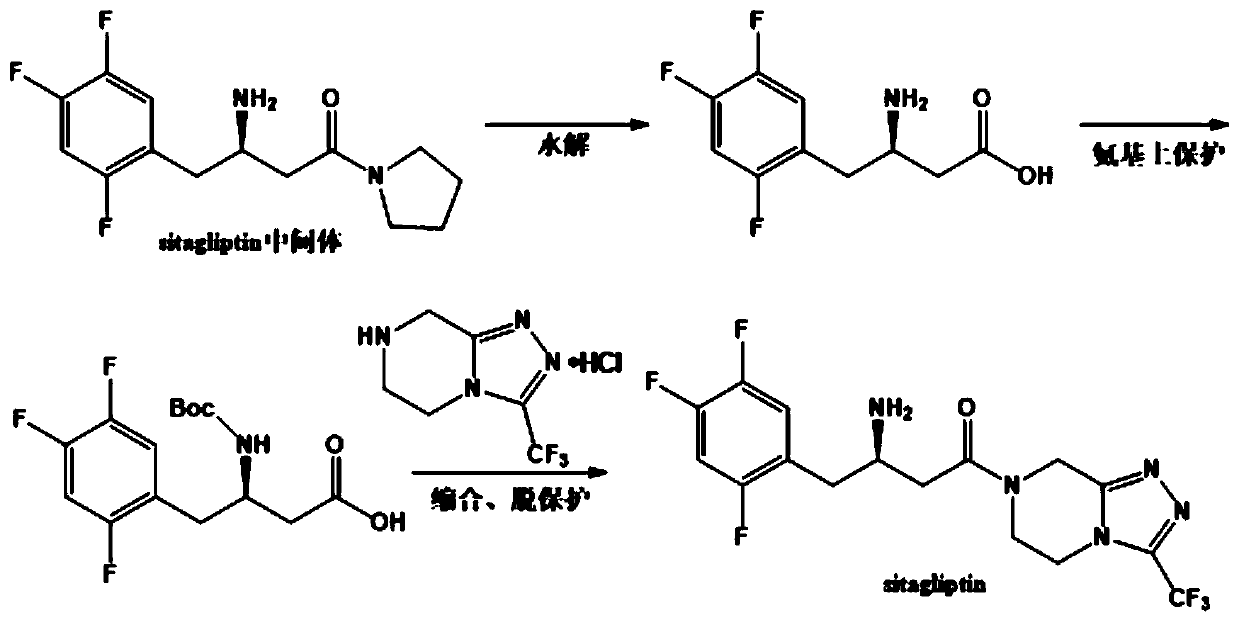
(R)-omega-transaminase mutant and application thereof in preparation of sitagliptin intermediate
ActiveCN111534494AIncrease enzyme activityIncreased substrate toleranceBacteriaTransferasesArginineThreonine
The invention discloses a (R)-omega-transaminase mutant and an application thereof in preparation of a sitagliptin intermediate. The mutant is obtained through multipoint mutation of arginine at the 77th site, leucine at the 181st site, arginine at the 130th site, tyrosine at the 139th site and threonine at the 273th site of an amino acid sequence shown as SEQ ID NO.1. According to the invention,a novel (R)-omega-TA recombinase is screened through a gene mining technology; molecular modification is carried out through a protein engineering technology; an (R)-omega-TA mutant catalyst with highenzyme activity, high substrate tolerance and high stereoselectivity is obtained; and the mutant can be used for asymmetric catalytic synthesis of the sitagliptin intermediate (R)-3-amino-1-(pyrrolidine-1-yl)-4-(2, 4, 5-trifluorophenyl) butan-1-one by taking a precursor ketone analogue 1-(pyrrolidine-1-yl)-4-(2, 4, 5-trifluorophenyl)-1, 3-butanedione as a substrate, and has higher conversion rate.
Owner:ZHEJIANG UNIV OF TECH +2
Collagenase producing strain as well as collagenase gene sequence and application thereof
InactiveCN106957803AInsensitiveIncrease enzyme activityCosmetic preparationsBacteriaHeterologousEscherichia coli
The invention relates to a collagenase producing strain, namely bacillus cereus, heterologous expression of a collagenase gene thereof and application thereof. The strain is separated and sieved from a soil sample and belongs to bacillus cereus after being identified. The strain is capable of extracellularly producing protease with collagen hydrolytic activity, and the protease with a molecular weight of 100kDa and collagenase activity is obtained by separating and purifying thallus fermentation supernate. Meanwhile, the collagenase gene of the strain is obtained through gene mining, and a collagenase protein with a molecular weight of 100kDa is obtained through heterologous expression and purification of escherichia coli. Researches confirm that the collagenase obtained through fermentation and purification of the Col15 strain and the collagenase obtained through heterologous expression and purification of escherichia coli are the same protein containing signal peptide. The collagenase produced by bacillus cereus Col15 has the properties of relatively wide temperature tolerance and pH value tolerance range and the like. Products obtained by hydrolyzing collagen and analogues thereof by virtue of the strain or the collagenase have wide economic prospects in the industries of foods, cosmetics, medical treatment and the like.
Owner:TIANJIN INST OF IND BIOTECH CHINESE ACADEMY OF SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com