Tumor mutation site screening and mutual exclusion gene mining method
A technology of mutation sites and mutated genes, which is applied in the field of tumor mutation site screening and mutually exclusive gene mining, and can solve problems such as unintuitive screening conditions.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0035] It should be noted that, in the case of no conflict, the embodiments of the present invention and the features in the embodiments can be combined with each other.
[0036] The present invention will be described in detail below with reference to the accompanying drawings and examples.
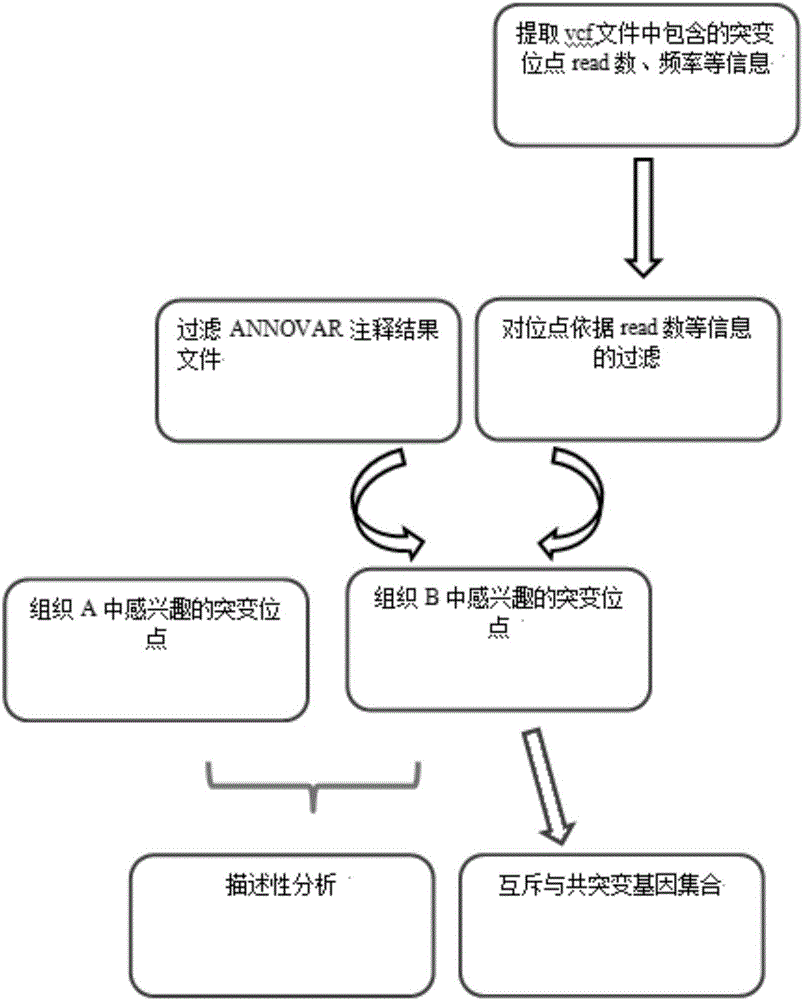
[0037] Method principle of the present invention is as follows:
[0038] The occurrence of gene mutation is a prerequisite for tumorigenesis, and the identification of mutated genes (gene loci) through exome sequencing data is one of the important means of biomedical research. The genes that have a greater impact on the physiological state of the cell are those genes that have a certain impact on the protein translated by the gene. Therefore, in order to identify tumor-related mutation genes (gene loci), we first filter the protein function on the annotation files generated by the commonly used annotation software ANNOVAR .
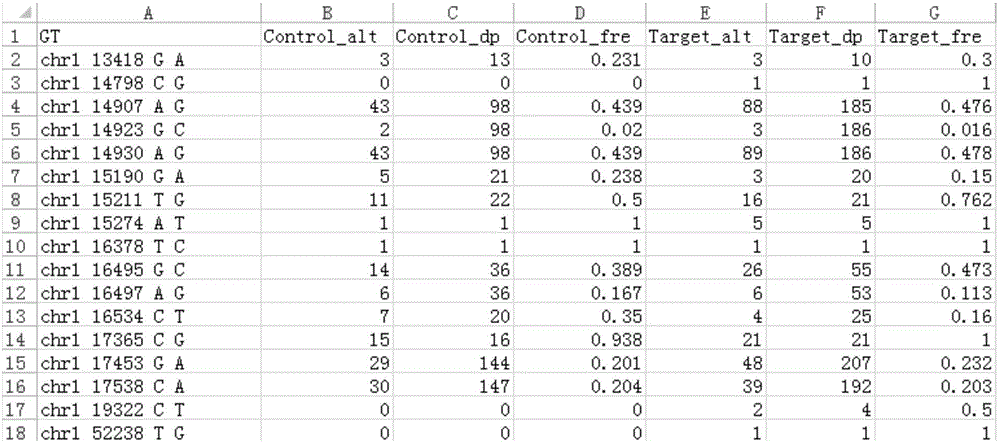
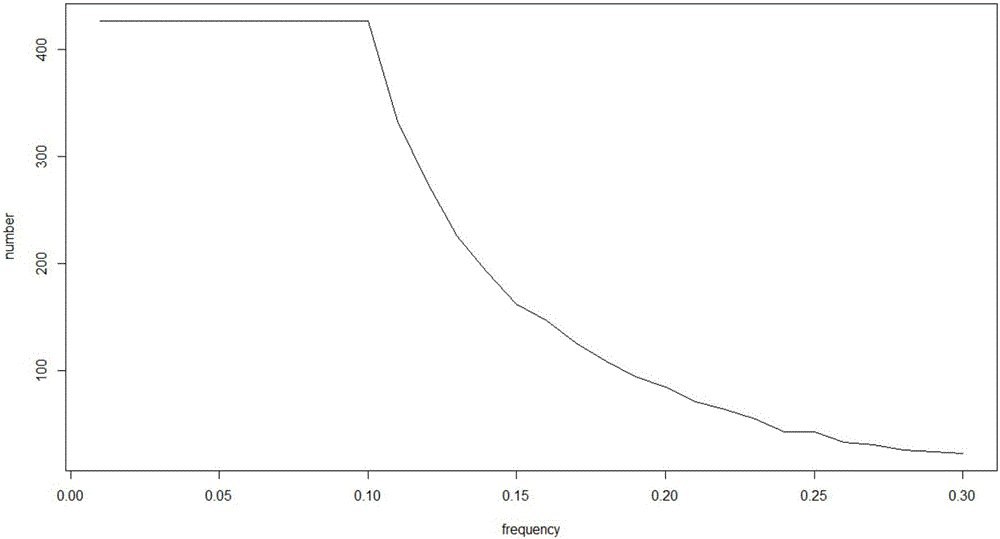
[0039] Next, due to the possibility of sequencing errors in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com