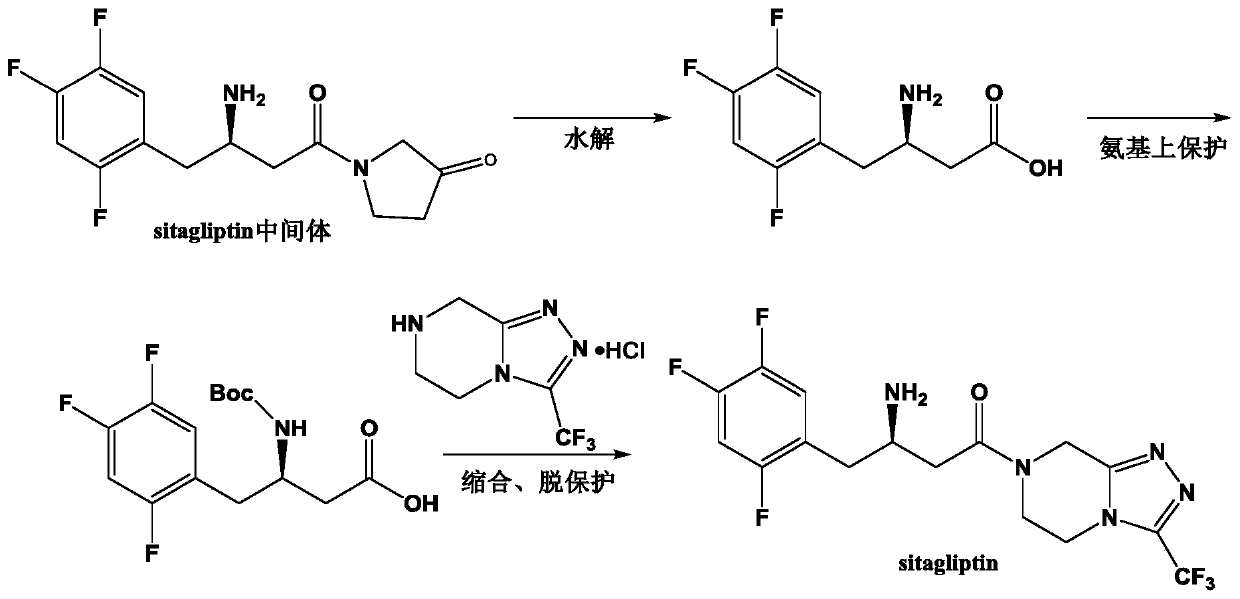
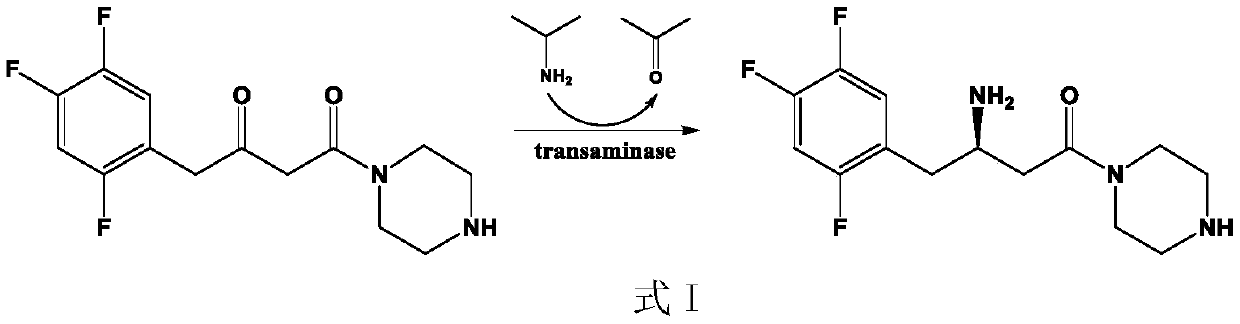
(R)-omega-transaminase mutant and application thereof
A technology of mutants and transaminases, applied in applications, transferases, enzymes, etc., can solve problems such as lack of competitive advantages, and achieve the effects of high stereoselectivity, high substrate tolerance, and high enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
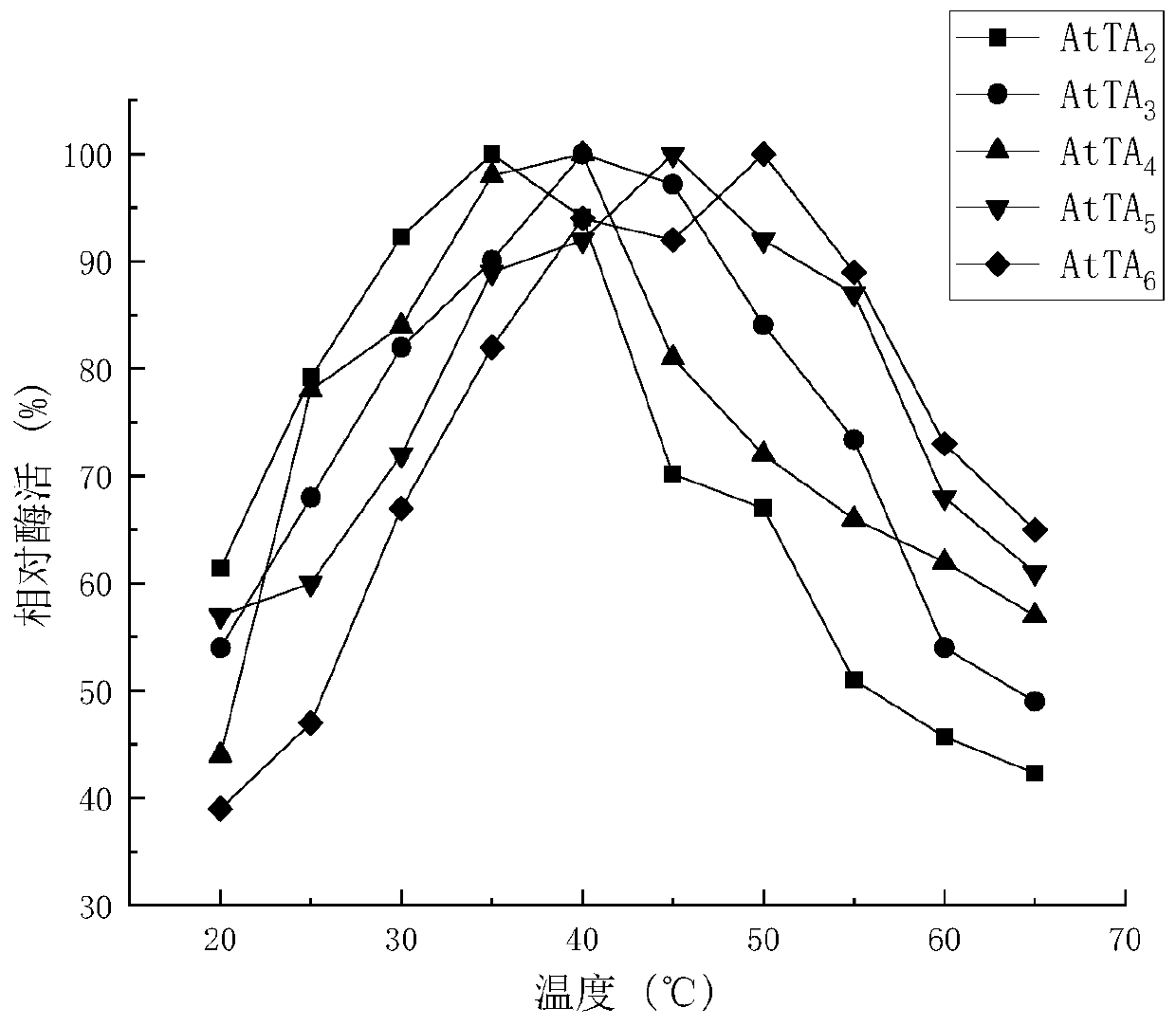
[0032] Example 1 Screening, determination of stereoselectivity and precise determination of enzyme activity of novel ω-TA
[0033] 1. Enzyme source and gene synthesis
[0034] Using the amino acid sequence of the commercial enzyme (R)-ω-TA 117 as a template, three ω-TA strains were obtained by gene mining from the NCBI database, namely Aspergillus terreus TA (AtTA, GenBank No. XP_001209325.1), Mycolicibacterium wolinskyi TA (MwTA , GenBank accession WP_067853383) and Aspergillus parasiticus TA (ApTA, GenBank accession KJK66446).
[0035] The sequence identity rates of the above three enzymes with (R)-ω-TA 117 were 38.14%, 48.97% and 37.9%, respectively. Codon optimization was carried out according to the codon preference of E.coli, and three strains of enzymes were synthesized by the method of whole gene synthesis, 6×His-tag tags were added at the end of the nucleic acid sequence, and restriction sites Xho I and Nco I were added at both ends, The gene was cloned into the cor...
Embodiment 2
[0058] Example 2 Construction and screening of AtTA single point mutants
[0059] 1. Mutant construction
[0060] The novel (R)-ω-TA screened was subjected to single-point mutation, and mutation primers were designed according to the nucleotide sequence of AtTA (shown as SEQ ID NO.2), and the recombinant vector pET28b / AtTA was used as the recombinant vector by using rapid PCR technology. Template, introduce a single mutation at position 182 of the amino acid sequence of AtTA, and the primers are:
[0061] Forward primer: TGAAAAAT NNK CAGTGGGGTGATCTGGTGC (the underline is the mutated base, as shown in SEQ ID NO.4)
[0062] Reverse primer: CCCCACTG MNN ATTTTTCACAGTCGGGTCA (the underline is the mutant base, as shown in SEQ ID NO.5)
[0063] PCR reaction system: 2×Phanta Max Buffer (containing Mg 2+ ) 25μL, dNTPs 10mM, forward primer 2μL, reverse primer 2μL, template DNA 1μL, Phanta Max Super-Fidelity DNA Polymerase 50U, add ddH 2 0 to 50 μL.
[0064] The PCR amplificatio...
Embodiment 3
[0078] Example 3 Construction and screening of AtTA two-site mutants
[0079] The single mutant AtTA constructed according to Example 2 1 Sequence design of mutation primers for site-directed mutagenesis, using rapid PCR technology to recombinant vector pET28b / AtTA 1 as the template, the AtTA 1 A single mutation is introduced at position 79 of the amino acid sequence, and the primers are:
[0080] Forward primer: ACATTACG NNK CTGGAGGCTAGCTGCACCA (the underline is the mutant base, as shown in SEQ ID NO.6)
[0081] Reverse primer: GCCTCCAG MNN CGTAATGTGGTCATCCAGA (the underline is the mutant base, as shown in SEQ ID NO.7)
[0082] The PCR reaction system is the same as the "construction of mutants" in Part 1 of Example 2.
[0083] The PCR amplification conditions were: 95°C for 3min; (95°C for 15s, 50°C for 15s, 63°C for 6.5min) 30 cycles; 72°C for 5min.
[0084] The PCR product was transformed into E.coli BL21(DE3) competent cells, and a single clone was picked in LB l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com