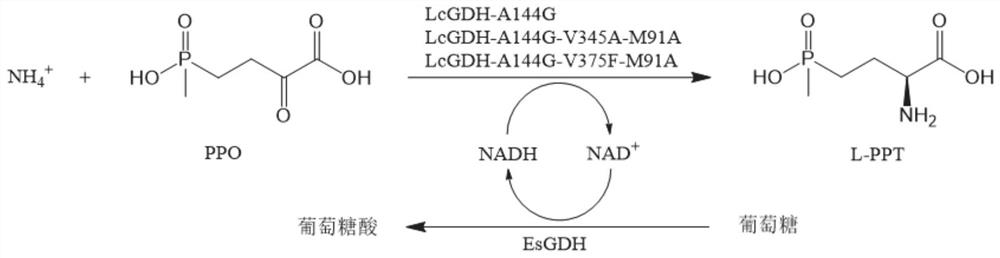
Gene mining method combining functional sequence and structural simulation, NADH preference type glufosinate-ammonium dehydrogenase mutant and application
A technology of glufosinate-ammonium and dehydrogenase, which is applied in genomics, instrumentation, proteomics, etc., can solve the problem of low activity of asymmetric amination reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Step 1: Analyze the characteristic sequences that NADH-type glutamate dehydrogenase should possess: ① protein size: the length of the candidate protein (300-500 amino acids), ② two necessary characteristic sequences of glufosinate-ammonium dehydrogenase: the first The segment is GGGKGG, where the X position represents any amino acid, and the second segment is one of VVTG, FVTG, VLTG, VFTG, FITG, FFTG, VVFG, FVFTG, VLFG, VFFG, FLFG, FFFG, ③The characteristic sequence of NADH binding : GXRVXXG.
[0048] Step 2: Search the gene bank: use the above characteristic sequences to perform iterative PSI-BLAST search and cluster analysis on the NCBI microbial genome resources and then the NCBINR sequence database (containing about 100 million protein genes), and obtain 15 clusters, 15 The degrees of aggregation of the clusters are 0.82, 0.76, 0.71, 0.66, 0.65, 0.58, 0.43, 0.42, 0.40, 0.39, 0.38, 0.34, 0.33, 0.32, 0.30 (arranged from high to low).
[0049] Step 3: Select represent...
Embodiment 2
[0050] Example 2: Construction and screening of glufosinate-ammonium dehydrogenase mutant library
[0051] The codon-optimized LcGDH amino acid sequence of Example 1 (the codon-optimized nucleotide sequence is shown in SEQ ID No.1), and the LcGDH gene obtained by gene synthesis by Hangzhou Qingke Biotechnology Co., Ltd. was cloned into On the NcoI of MCS1 (Multiple Cloning Site 1) of the plasmid pETDuet, construct the recombinant expression vector pETDuet-LcGDH, retain the His-Tag gene of the plasmid itself, transform it into E. coli BL21 (DE3), and send it to Hangzhou Qingke Biotechnology Co., Ltd. synthesized wild-type glufosinate-ammonium dehydrogenase engineering bacteria E.coli BL21(DE3) / pETDuet-LcGDH.
[0052] The glucose dehydrogenase gene EsGDH was cloned from Exiguobacterium sibiricum ZJBML01011 (the nucleotide sequence is shown in GenBank accession number KM817194.1), and constructed into the recombinant expression vector pETDuet-LcGDH by Vazyme's OneStep Cloning Kit...
Embodiment 3
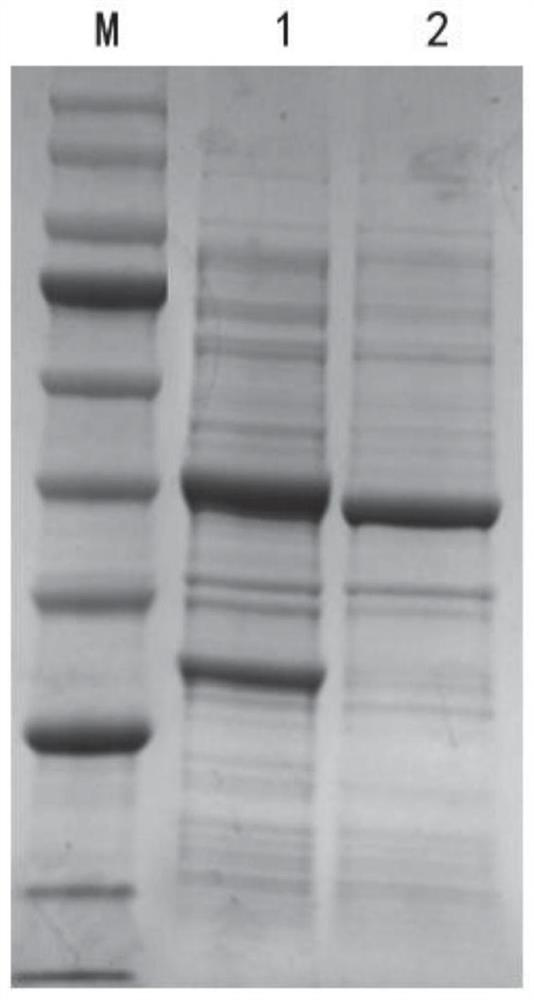
[0060] Embodiment 3: Induced expression of glufosinate-ammonium dehydrogenase mutant engineering bacteria
[0061] The wild-type glufosinate-ammonium dehydrogenase and glucose dehydrogenase in Example 2 are set out to co-express strain E.coliBL21(DE3) / pETDuet-1-LcGDH-EsGDH and the glufosinate-ammonium dehydrogenase mutant with glucose dehydrogenase Enzyme co-expression strains:
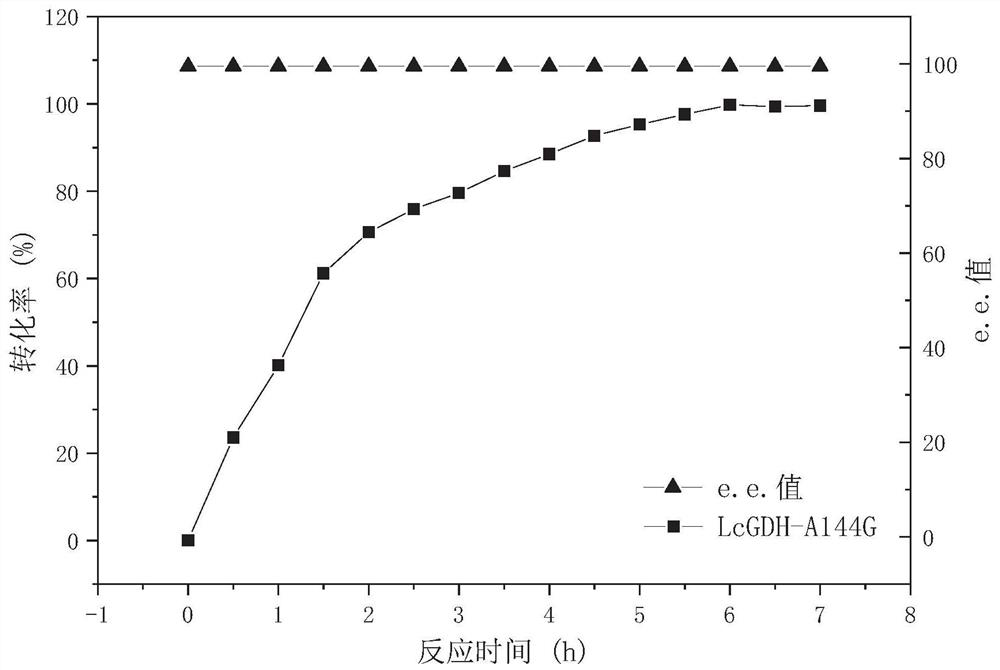
[0062] E.coli BL21(DE3) / pETDuet-1-LcGDH(A144G)-EsGDH,
[0063] E.coli BL21(DE3) / pETDuet-1-LcGDH(A144G-V345A-M91A)-EsGDH,
[0064] E.coli BL21(DE3) / pETDuet-1-LcGDH(A144G-V375F-M91A)-EsGDH,
[0065] Inoculate them into LB liquid medium containing a final concentration of 50 μg / mL ampicillin, culture at 37°C for 8 hours, and inoculate them into fresh LB liquid culture medium containing a final concentration of 50 μg / mL ampicillin at an inoculum volume concentration of 2%. culture medium at 37°C and 180 rpm for 2 hours, and then add 0.1mM IPTG to the culture medium at a final concentration of 18°C fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com