Soil microbe genome DNA extracting method
A technology of soil microorganisms and extraction methods, applied in the direction of recombinant DNA technology, biochemical equipment and methods, DNA preparation, etc., can solve the problems of pollution and high purity, and achieve the effects of cost reduction, high purity and wide application range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
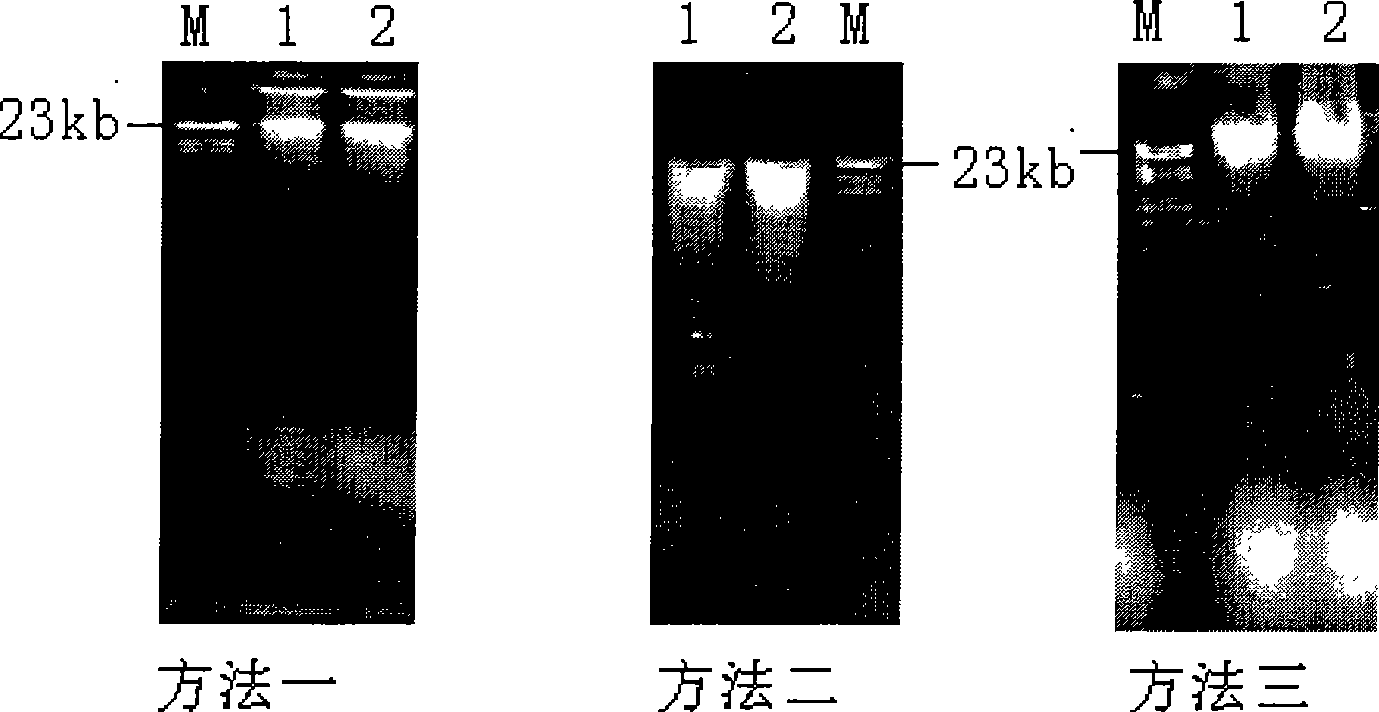
[0023] Taking the black soil of farmland in Northeast China as an example (see Table 2), three methods were used to extract the genomic DNA of soil microorganisms:
[0024] Table 2 Physical and chemical properties of tested soil / g·kg -1
[0025]
[0026] Method one is the method provided by the invention:
[0027] 1) Preliminary lysing of microbial cells: Take 0.5g of soil and 0.5g of quartz sand and put them into a 1.5ml screw cap centrifuge tube, add 850μl of extraction buffer, and shake for 40S with a nucleic acid extractor (Q-biogene, USA, model FP220) at a speed of 40 seconds. It is 5.5m / s.
[0028] 2) Further lysis of microbial cells: 50 μl of lysozyme with a concentration of 100 mg / ml was added to the initially lyzed cell solution and mixed evenly. After incubating at 37° C. for 30 minutes, 100 μl of 20% (w / v) SDS was added, mixed evenly, and then Incubate at 65°C for 30 minutes, then centrifuge at 12,000 rpm for 5 minutes at room temperature, and collect the supe...
Embodiment 2
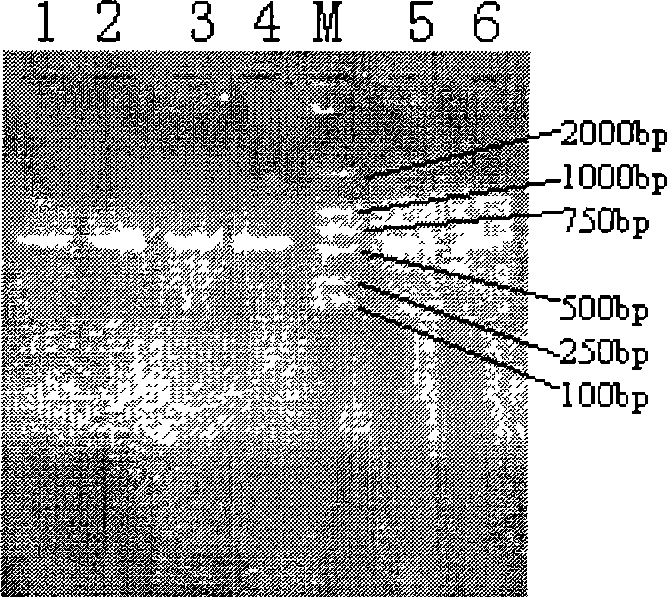
[0038] The genomic DNA extracted by the three methods in Example 1 was used as a template to amplify a partial fragment of the 16S rRNA gene, and the primers for PCR amplification were 27F:5'-GAGAGTTTGATCCTGGCTCAG-3'519R:5'-ATTACCGCGGCTGCTGG-3', in the preceding A GC clamp was added to the 5′ end of the primer: 5′-cgcccgccgcgcgcggcgggcggggcgggggcccggggg-3′. The PCR reaction system is: buffer solution: 100mM Tris, 100mM EDTA and 1.5M NaCl, pH8.0; MgCl 2 The final reaction concentration is 2.0 mM; the final reaction concentration of dNTP is 0.2 mM; the final reaction concentration of the front primer P1 is 0.5 μM; the final reaction concentration of the rear primer P2 is 0.5 μM; Taq enzyme (Takara Company) 1 U;
[0039] PCR reaction conditions: pre-denaturation: 94°C, 5min; denaturation: 94°C, 10s, annealing: 63°C, 30s, 10 cycles, each cycle annealing temperature decreased by 1°C. Extension: 68°C, 2min; denaturation: 94°C, 2min, annealing: 53°C, 30s, 25 cycles; extension: 68°C,...
Embodiment 3
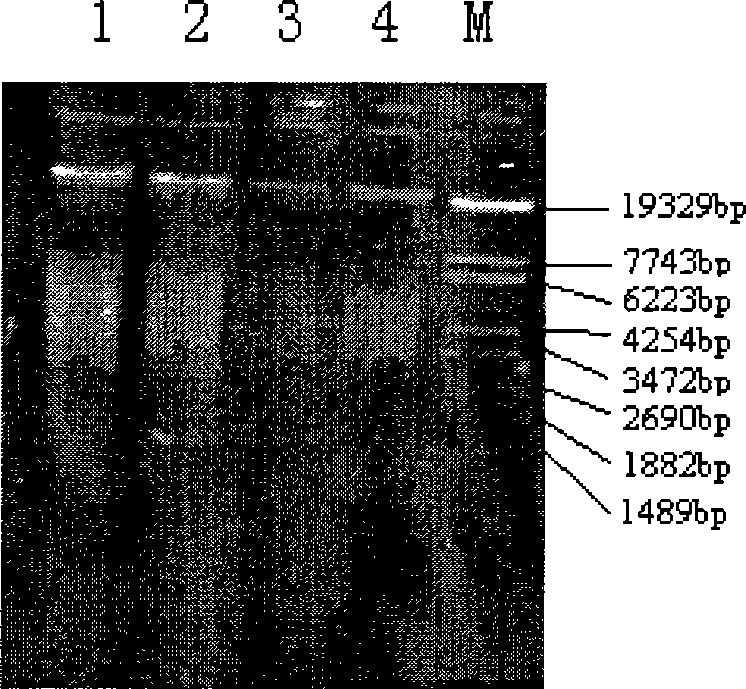
[0042] Adopt the method of the present invention to extract four kinds of soils simultaneously: paddy field soil, upland umber soil, straw mat soil, upland black soil, the first three kinds of soil samples are collected to the Shenyang Ecological Station of the Shenyang Institute of Ecology, Chinese Academy of Sciences, and the black soil is collected to the Heilongjiang Agricultural Ecological Experiment of the Chinese Academy of Sciences stand. The extracted DNA was subjected to agarose gel electrophoresis, the gel concentration was 6%, the voltage was 50V, and the electrophoresis was performed for 2 hours, and the concentration and purity of the extracted nucleic acid were detected simultaneously (see Table 4).
[0043] Table 4. Four kinds of soil genomic DNA extraction results
[0044] sample Paddy soil Upland Brown Soil bedding soil Dry field black soil DNA yield (μg / ml) 107.5 84.2 62.5 150.1 OD 260nm / OD 280nm 1.84 1.88 1.99 1.78 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com