Process for prodn. of L-Thr
A technology of threonine and antibiotics, applied in the field of producing L-threonine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1: Constructing a recombinant plasmid and using it to knock out the tdc gene
[0037] Genomic DNA of the threonine-producing strain pGmTN-PPC (KCCM-10236) was isolated using the QIAGEN Genomic-tip system. Using genomic DNA as a template, a 3.1kb tdc operon gene fragment (5295pb) containing tdc B and tdc C was amplified by polymerase chain reaction (PCR). Primers used were 5'-agg agg gga tcc ggt atg tct tct gag gcg-3' and 5'-agg agg gaa ttc atc ggc aac agg cac ag-3'. PCR was performed as follows, 30 cycles of amplification, each cycle including denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 3 minutes and 30 seconds.
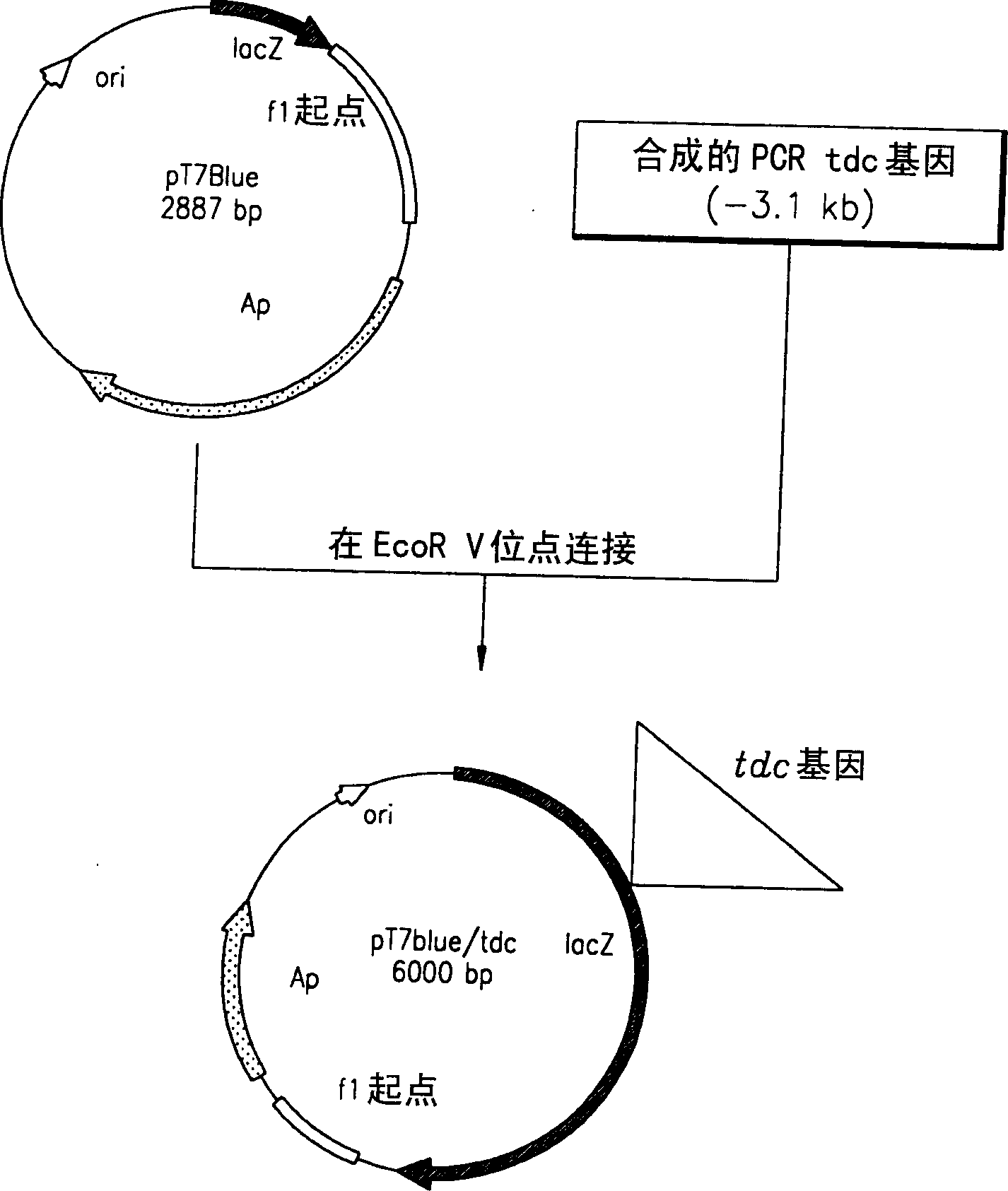
[0038] The PCR product was electrophoresed on a 0.7% agarose gel, and the band of desired size was eluted. Elution band and pT7Blue cloning carrier (Novagen Co,) blunt-end ligated overnight in order to obtain recombinant plasmid pT7Blue / tdc at 16 ℃ (referring to figure 1 ). Escherichia coli DH5α...
Embodiment 2
[0040] Example 2: Screening for strains integrated with recombinant plasmids
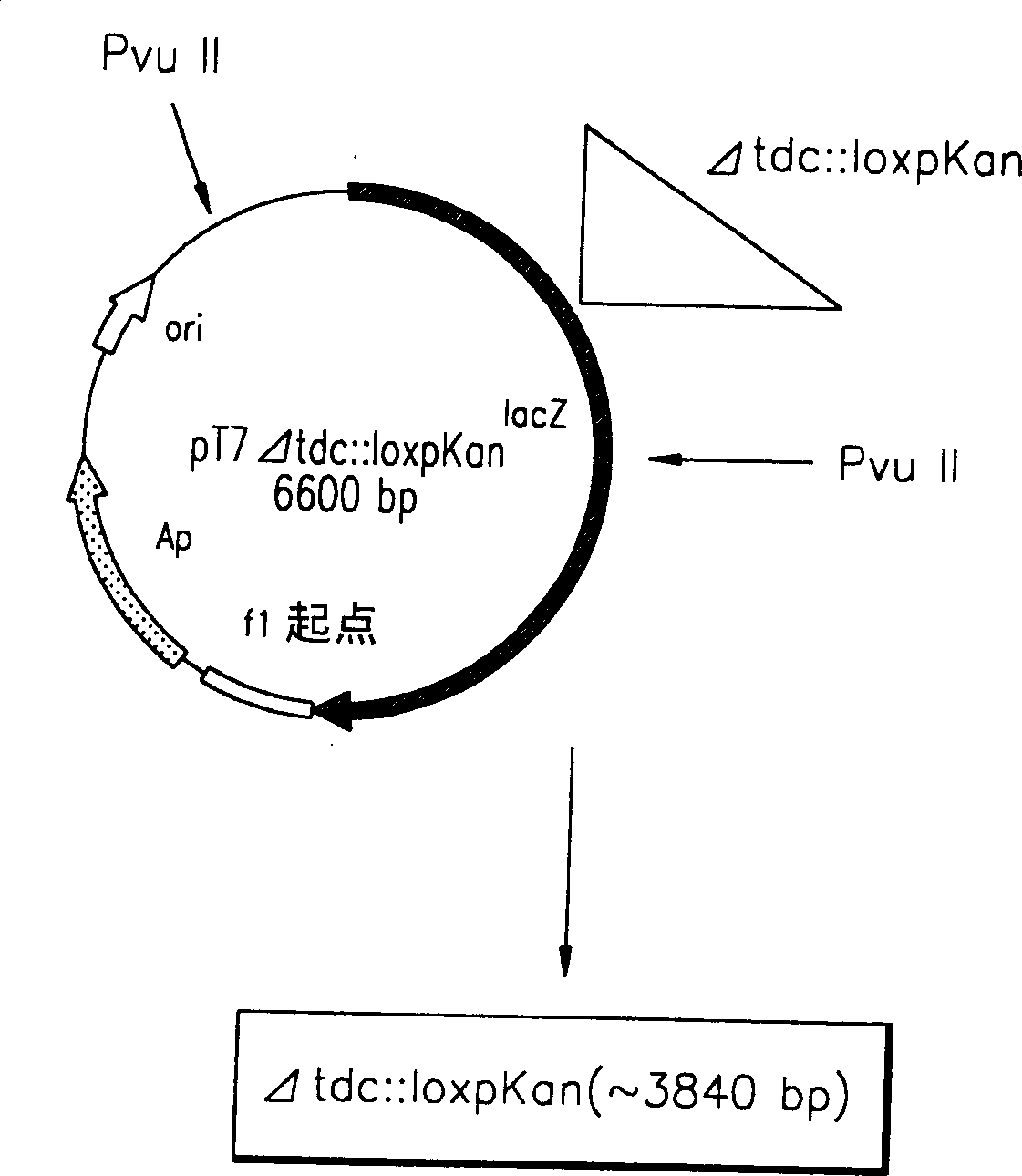
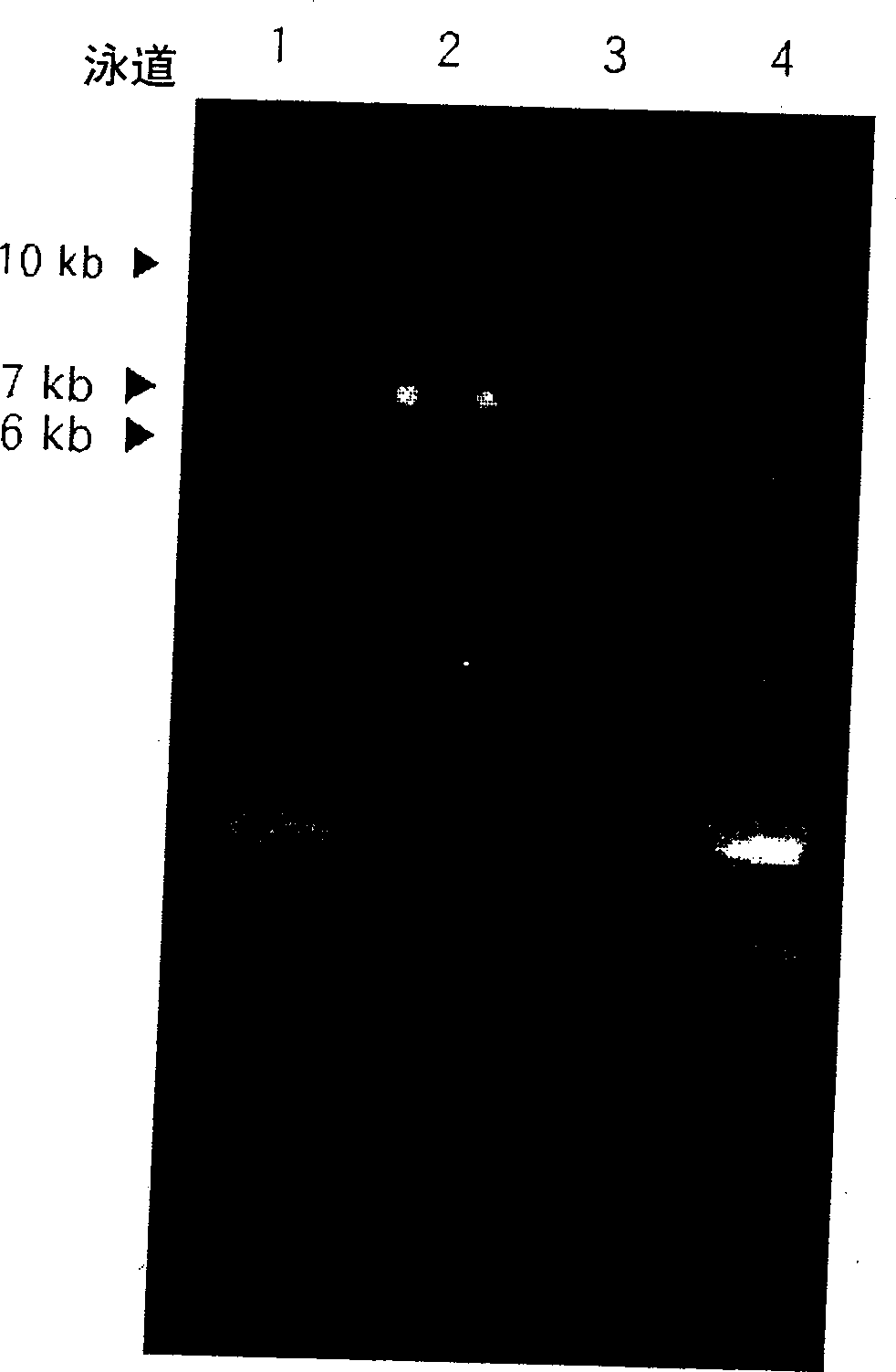
[0041] Escherichia coli DH5α was transformed with the recombinant plasmid pT7Δtdc::loxpKan, plated on a solid medium containing 50 mg / L ampicillin and 15 mg / L kanamycin, and cultured overnight at 37°C. Pick colonies from the culture with a toothpick, inoculate into 3 ml of liquid medium containing ampicillin and kanamycin, and cultivate overnight at 200 rpm. Plasmid DNA was isolated from the cultures using a QIAGEN miniprep kit, and the size of the plasmid DNA was identified. Plasmid DNA was digested with restriction enzymes and loaded onto 0.7% agarose to confirm its orientation. The identified plasmid DNA was digested with restriction enzyme PVU II and electrophoresed on a 0.7% agarose gel to elute a DNA fragment of about 3840 kb (Δtdc::loxpKan). The threonine-producing strain pGmTN-PPC12 was transformed with the DNA fragment Δtdc::loxpKan by electroporation, and plated on a solid medium conta...
Embodiment 3
[0042] Embodiment 3: the shake flask culture comparison threonine output of the recombinant bacterial strain that obtains
[0043] The L-threonine production comparison of 30 single colonies of the recombinant strains cultured in the kanamycin-containing solid medium in Example 2 was screened in an Erlenmeyer flask (flask) using a threonine titer medium. The composition of the threonine titer medium used in each case is shown in Table 1.
[0044] components
content per liter
glucose
70g
(NH 4 ) 2 SO 4
28g
K H 2 PO 4
1.0g
MgSO 4 ·7H 2 o
0.5g
FeSO 4 ·7H 2 o
5mg
MnSO 4 ·8H 2 o
5mg
30g
0.15g
yeast extract
2g
PH7.0
[0045] Single colonies were grown overnight at 32°C on LB solid medium in an incubator. One full loop of each culture was inoculated into 25 ml of titer medium a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com