Extraction method of microbe DNA for high-throughput sequencing in sample
An extraction method and high-throughput technology, which is applied in the fields of high-throughput gene sequencing and DNA extraction, can solve problems such as difficulties, low cell lysis rate, and inaccessibility, and achieve fast extraction speed, low degradation degree, and good versatility Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1 The present invention is used for the preparation of the reagent of extracting microbial DNA
[0037] The reagent consumables for extracting microbial DNA according to the present invention include:
[0038] Lysis buffer contains: 100mM Tris-HCl, 100mM EDTA, 100mM Na 3 PO 4 , 1.5M NaCl, and 0.1% CTAB. Specific configuration method: weigh 0.82gNa respectively 3 PO 4 and 0.5gCTAB, add appropriate amount of distilled water to dissolve respectively, then add 5mL of 1M Tris-HCl, 10mL of 0.5M EDTA, 15mL of 5M NaCl, and then make the volume to 50mL.
[0039] Aluminum ammonium sulfate solution: 50-200mM Al 2 (SO 4 ) 3 and 50-200mM (NH 4 ) 2 SO 4 , the two are mixed according to the volume ratio of 1:1. The aluminum ammonium sulfate solution in the embodiment of the present invention selects the Al of 120mM for use 2 (SO 4 ) 3 and 120mM (NH 4 ) 2 SO 4 It is obtained by mixing in a volume ratio of 1:1.
[0040] Binding solution: 5M guanidine hydroch...
Embodiment 2
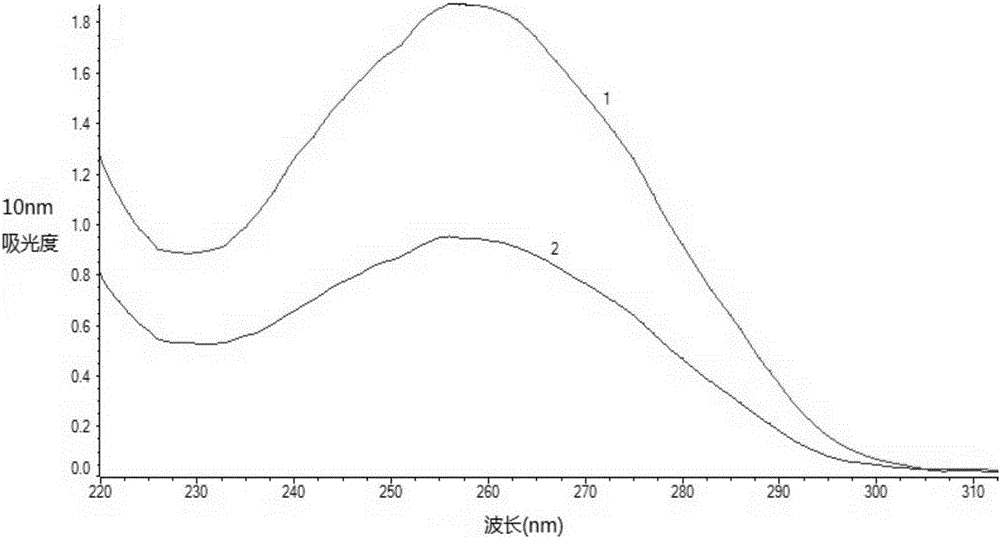
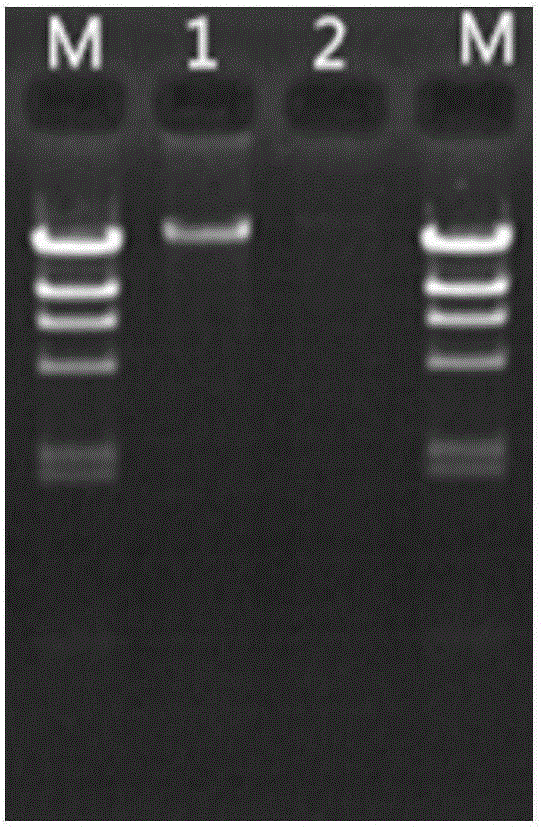
[0044] Example 2 Microbial DNA Extraction and Effect Verification from Soil
[0045] The reagents and consumables described in Example 1 were used for extraction.
[0046] (1) the method for extracting microbial DNA in the soil of the present invention
[0047] 1. Weigh about 500mg of soil sample into a 2mL centrifuge tube, add 1.25mL of lysis buffer, add 25uL of β-mercaptoethanol at the same time, and vortex for 2 minutes. Then 30 μL of lysozyme (15 mg / mL) was added and placed on a shaker at 37° C. for 15 min.
[0048] 2. Add 250 μL of 10% SDS and 20 μL of proteinase K (20 mg / mL) solution, and immediately vortex to mix thoroughly. Water bath at 65°C for 10 minutes, and mix by inverting every 2 minutes during this period.
[0049] 3. Centrifuge at 10000g for 2min at room temperature, and transfer the supernatant to a clean 1.5mL centrifuge tube.
[0050] 4. Add 1 / 3 volume of 5M NaCl solution to the supernatant, vortex and mix for 5 seconds, and place in an ice bath at 4°C ...
Embodiment 3
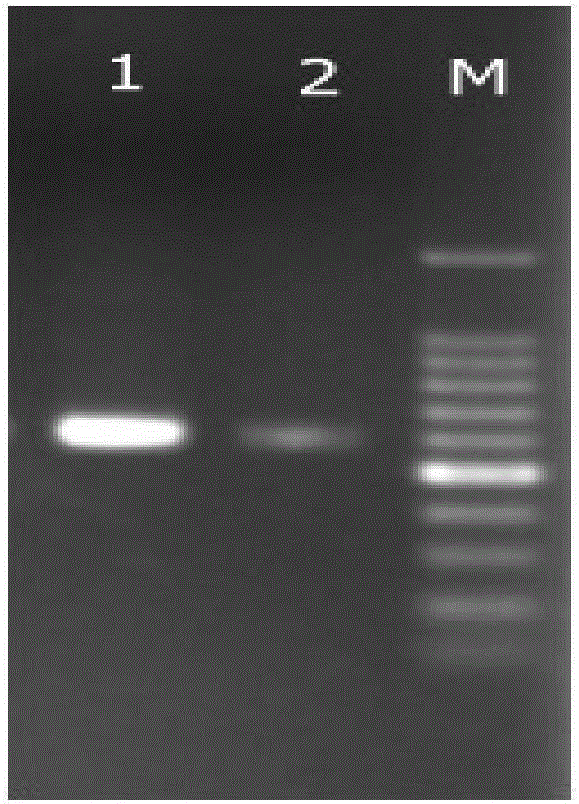
[0061] Example 3 Microbial DNA Extraction and Effect Verification from Feces
[0062] The reagents and consumables described in Example 1 were used for extraction.
[0063] The method for extracting microbial DNA in feces of the present invention:
[0064] 1. Weigh about 500mg of cryopreserved cow manure sample into a 2mL centrifuge tube, add 25uL β-mercaptoethanol and 1.25mL lysis buffer, and vortex for 2 minutes. Then 30 μL of lysozyme (15 mg / mL) was added and placed on a shaker at 37° C. for 15 min.
[0065] 2. Add 250 μL of 10% SDS and 20 μL of proteinase K (20 mg / mL) solution, and immediately vortex to mix thoroughly. Water bath at 65°C for 10 minutes, and mix by inverting every 2 minutes during this period.
[0066] 3. Centrifuge at 10000g for 2min at room temperature, and transfer the supernatant to a clean 1.5mL centrifuge tube.
[0067] 4. Add 1 / 3 volume of 5M NaCl solution to the supernatant, vortex and mix for 5 seconds, and place in an ice bath at 4°C for 8 min...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com