Construction method of novel coronavirus whole-genome high-throughput sequencing library, and kit for library construction
A coronavirus and whole genome technology, applied in the biological field, can solve problems such as slow timeliness, high requirements, and low data volume requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
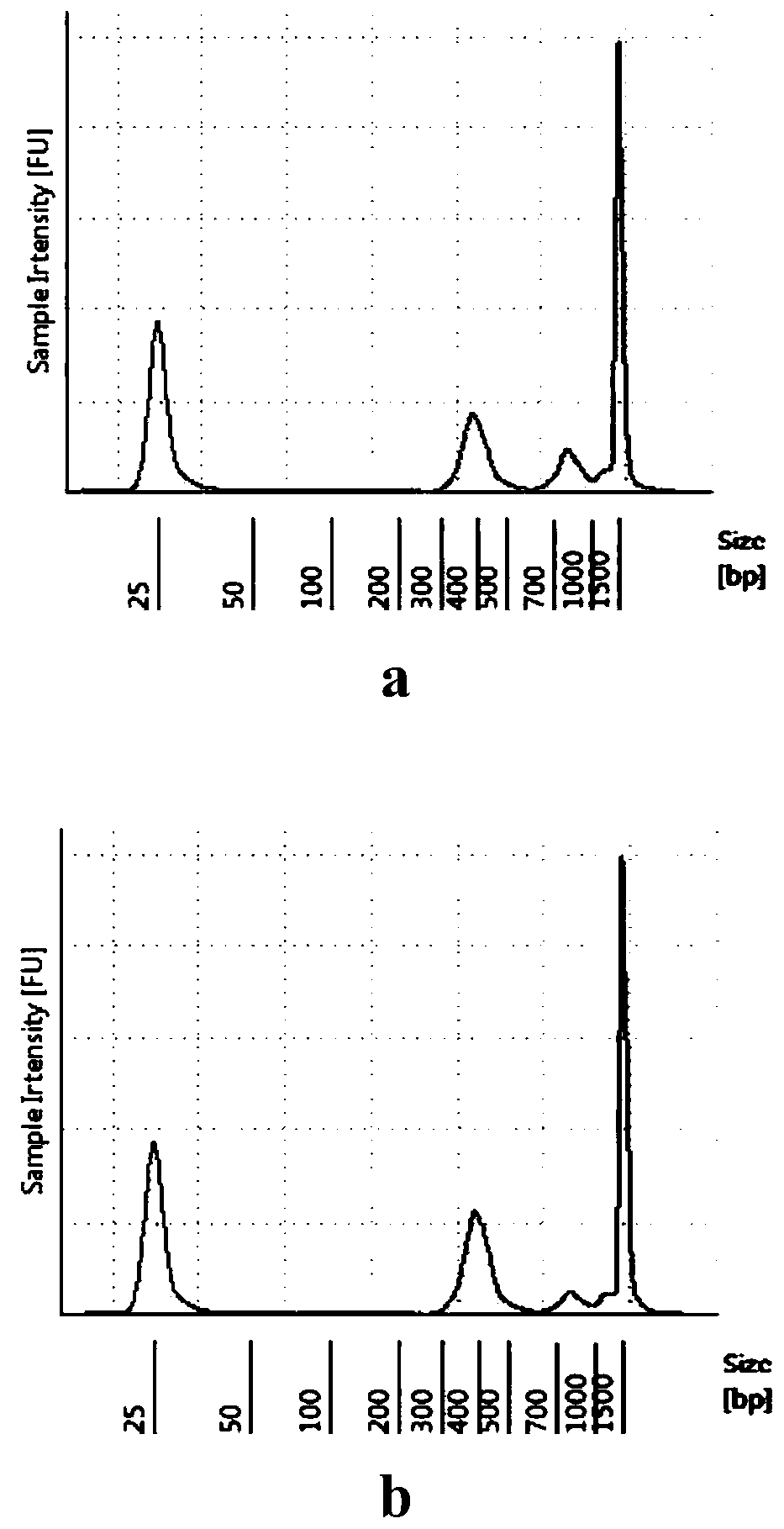
[0086] The viral RNA used in this example was extracted from the alveolar lavage fluid of patients with new coronary pneumonia by the magnetic bead method, a total of two cases; Safety level 3 (P3) laboratory completed.
[0087] The method provided in this example can be used to detect virus species in alveolar lavage fluid or to detect virus genome mutations from patients with confirmed new coronary pneumonia; the virus copy number is measured by the new coronavirus nucleic acid standard substance (high concentration) GBW (E) 091089 (China The N gene copy number of the Institute of Metrology and Science) was used as a reference, and the virus concentration (Copies / μL) was determined by absolute quantitative qRT-PCR (Table 3).
[0088] Table 3 RNA virus copy number and clinical information in alveolar lavage fluid
[0089]
[0090] The specific experimental method is as follows:
[0091] The viral single-stranded RNA extracted from the alveolar lavage fluid was reverse-tr...
Embodiment 2
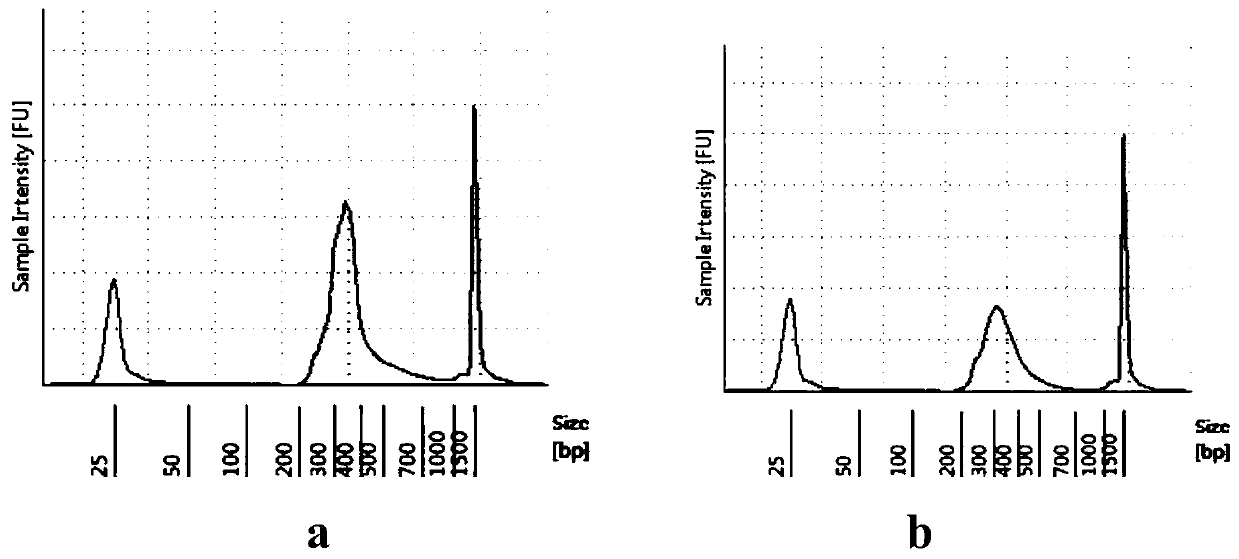
[0114] The viral RNA samples 46d1 and 50d1 used in this embodiment are the same as those in Embodiment 1.
[0115] The specific experimental method is as follows:
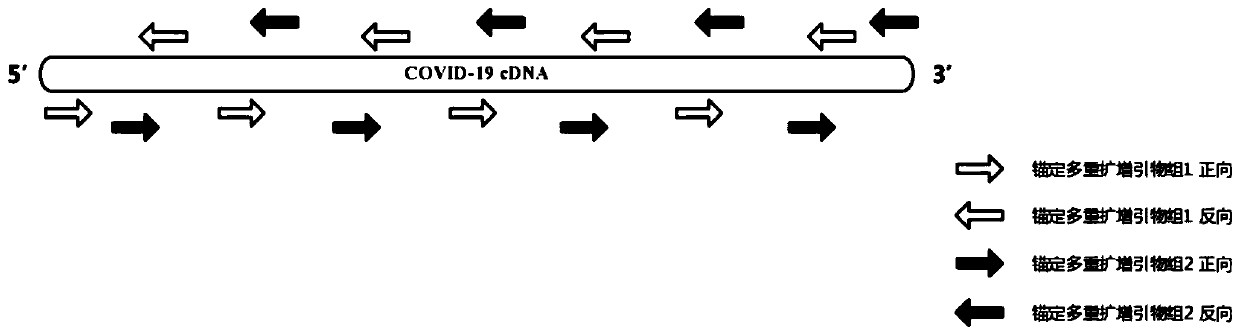
[0116] Viral single-stranded RNA extracted from alveolar lavage fluid used 34 gene-specific reverse primers (COV-1-R, COV-8-R, COV-12-R, COV-20-R, COV-30- R, COV-38-R, COV-47-R, COV-54-R, COV-62-R, COV-71-R, COV-80-R, COV-86-R, COV-94-R, COV-102-R, COV-111-R, COV-119-R, COV-125-R, COV-132-R, COV-141-R, COV-146-R, COV-155-R, COV- 162-R, COV-172-R, COV-179-R, COV-187-R, COV-195-R, COV-202-R, COV-210-R, COV-220-R, COV-228- R, COV-233-R, COV-239-R, COV-247-R, COV-252-R) mixed (genome direction 3'-5'), reverse transcribed into a strand of cDNA (1st cDNA), The 1st cDNA synthesis kit is selected as: TAKARA PrimeScript 1 st strand cDNA Synthesis Kit (TAKARA, CatNo.6110A); 1st cDNA was purified for subsequent amplification.
[0117] Use the anchored multiple amplification primer set 1 to carry out PCR reaction. The PC...
Embodiment 3
[0140] The viral RNA used in this example was extracted from throat swab samples of patients with new coronary pneumonia by magnetic bead method; the extraction and quality inspection of RNA was conducted by Chinese Academy of Medical Sciences / Peking Union Medical College Hospital Institute of Pathogen Biology Biosafety Level 3 ( P3) The laboratory is completed.
[0141] The method provided in this example can be used to detect virus species in throat swab samples or to detect viral genome mutations from patients with confirmed or suspected novel coronavirus pneumonia; The copy numbers of N gene and E gene (National Institute of Metrology, China) were used as reference, and the virus concentration (Copies / μL) was determined by absolute quantitative qRT-PCR (Table 8). .
[0142] Table 8 Pharyngeal swab RNA virus copy number, clinical information
[0143]
[0144] The specific experimental method is as follows:
[0145] The viral single-stranded RNA extracted by the magnet...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com