Method for detecting off-target effect of adenine base editor system based on whole genome sequencing, and its application in gene editing
A whole-genome sequencing and editing system technology, applied in the field of detection of off-target effects of adenine single base editing system, can solve problems restricting the application of ABE system and achieve the effect of wide application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0048] The present invention will be further described below in conjunction with the accompanying drawings and specific embodiments, but the embodiments do not limit the present invention in any form.
[0049] Unless otherwise specified, the reagents, methods and equipment used in the present invention are conventional reagents, methods and equipment in the technical field. Unless otherwise specified, the reagents and materials used in the following examples are commercially available. Experimental methods that do not indicate specific conditions are usually implemented under conventional conditions or conditions suggested by the manufacturer.
[0050] In a specific embodiment of the present invention, the present invention provides a system, method, kit and application thereof for detecting off-target effects of an adenine single base editing system based on whole genome sequencing.
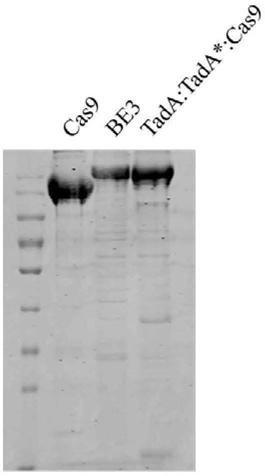
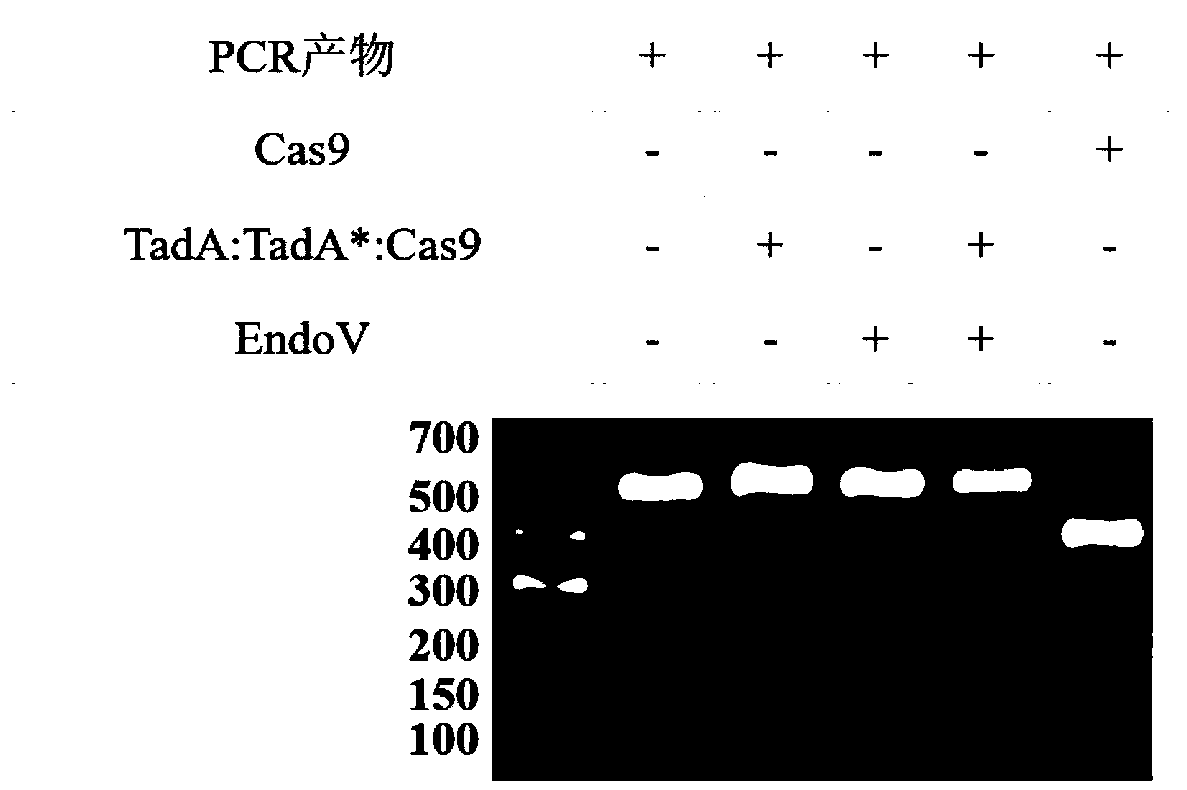
[0051] The method for detecting the off-target effect of the adenine single base editing sy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com