Eukaryotic gene editing method based on gene cas7-3 in I type CRISPR-Cas system
A gene editing and gene technology, applied in the field of genetic engineering and eukaryotic gene editing, to achieve the effect of effective gene editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
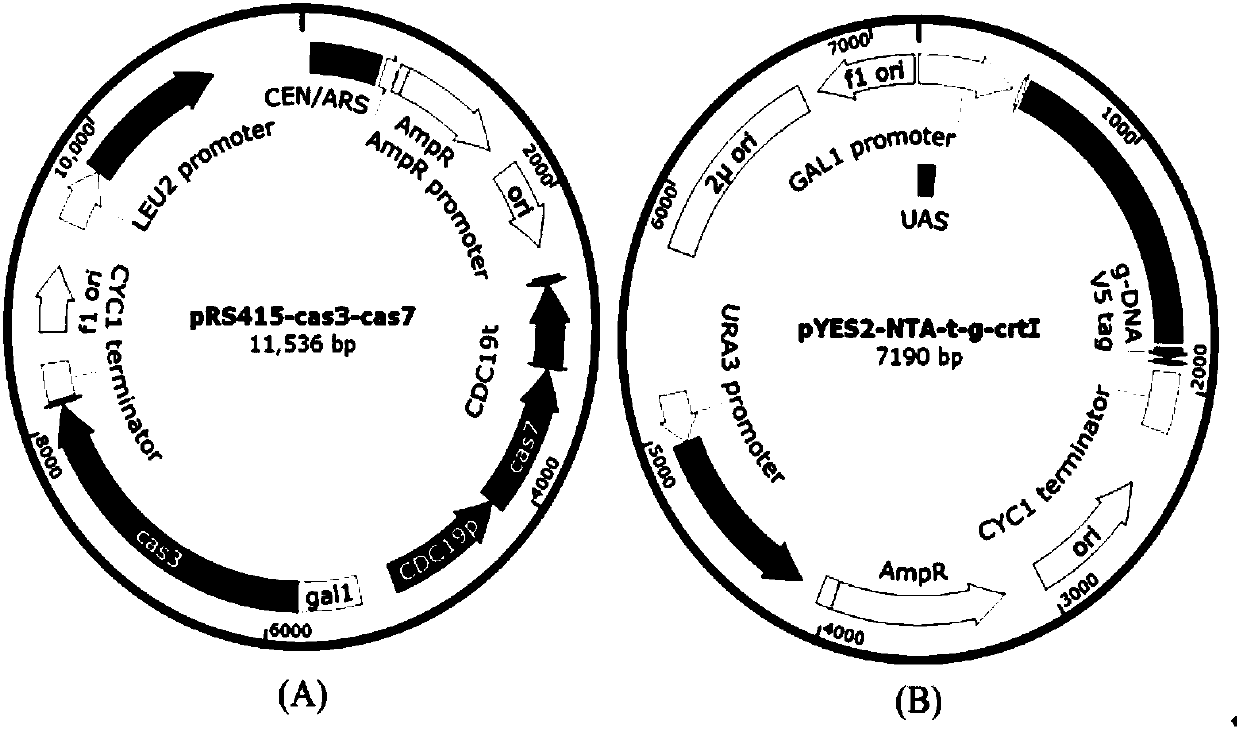
[0048] Example 1 pRS415-cas7-3 and pYES2 / NTA-t / g-ΔcrtI were respectively transformed to knock out the SC BY4741 gene crtI
[0049] (1) Construction of protein expression plasmid pRS415-cas7-3
[0050] (A) Construction of protein expression plasmid pRS415-cas3
[0051] According to the sequence information of plasmid pRS415, specific primers cas3-F / cas3-R and pRS415-cas3-F / pRS415-cas3-R were designed, and the two ends of the primers carried the complementary sequences of cas3 gene and plasmid pRS415-cas9 respectively. Cas3 gene PCR amplification was performed using TransStart FastPfu DNA Polymerase produced by Quanshijin Biotechnology Co., Ltd., and the reaction conditions were: 95±1°C for 5±1min, 95±1°C for 30s, 62±3°C for 30s, and 72°C for 1±1min ( 50μl reaction system), 30 cycles, 72°C for 10min. The PCR product was detected by 1% agarose electrophoresis, recovered by the kit, and a purified cas3 gene fragment was obtained. The same PCR amplification was performed again t...
Embodiment 2
[0073] Example 2 pRS415-cas7-3 and pYES2 / NTA-t-ΔcrtI were respectively transformed to knock out the SC BY4741 gene crtI
[0074] (1) Construction of protein expression plasmid pRS415-cas7-3
[0075] With embodiment 1 step (1)
[0076] (2) Construction of gene editing template vector pYES2 / NTA-t-ΔcrtI
[0077] (A) Preparation of upstream and downstream homology arms
[0078] With embodiment 1 step (2A)
[0079] (B) Construction of gene editing template vector pYES2 / NTA-t-ΔcrtI
[0080] With embodiment 1 step (2B)
[0081] (3) Acquisition and inspection of recombinants
[0082] (A) Competent preparation of SCBY4741 and transformation of pRS415-cas7-3
[0083] With embodiment 1 step (3-A)
[0084] (B) Competent preparation of SC BY4741-pRS415-cas7-3 and transformation of pYES2 / NTA-t-ΔcrtI
[0085] Except importing plasmid pYES2 / NTA-t-ΔcrtI, all the other are the same as embodiment 1 step (3B) (see figure 2 ).
Embodiment 3
[0086] Example 3 Transformation of single plasmid pRS415-cas7-3-t / g-ΔcrtI to knock out SC BY4741 gene crtI
[0087] (1) Construction of protein expression plasmid pRS415-cas7-3
[0088] With embodiment 1 step (1)
[0089] (2) Construction of protein expression and gene editing single plasmid pRS415-cas7-3-t / g-ΔcrtI
[0090] (A) Preparation of upstream and downstream homology arms
[0091] Except that crtI gene upstream homology arm primer crtI-UF2, downstream homology arm primer crtI-DR2 contain XhoI restriction enzyme site simultaneously, other is the same as embodiment 1 step (2A), used upstream homology arm primer crtI-UF2 The sequence of the primer crtI-DR2 is ccgCTCGAGaccaactgaacgagcaataacgg.
[0092] (B) Construction of protein expression and gene editing single plasmid pRS415-cas7-3-t / g-ΔcrtI
[0093] Except that the g-crtI-DNA and t-ΔcrtI-DNA fragments were respectively connected to the pRS415-cas7-3 plasmid, other steps were the same as step (2B) of Example 1.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com