THERAPEUTIC USES OF GENOME EDITING WITH CRISPR/Cas SYSTEMS
a genome editing and genome technology, applied in the field of therapeutic use of genome editing with crispr/cas systems, can solve the problems of only 2%-4% efficiency of allele targeting in human stem cells, and achieve the effect of effectively deleting target polynucleotide sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
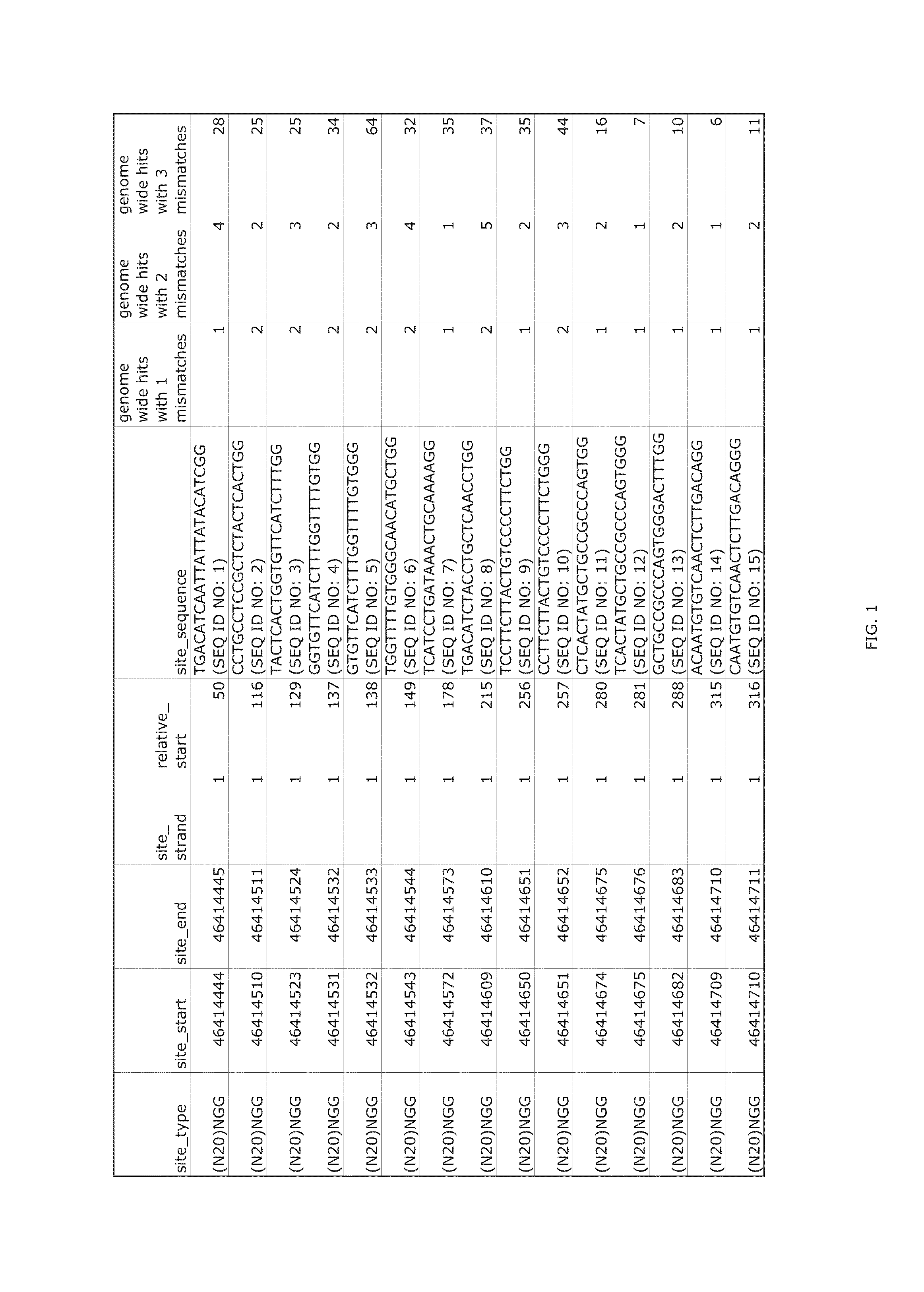
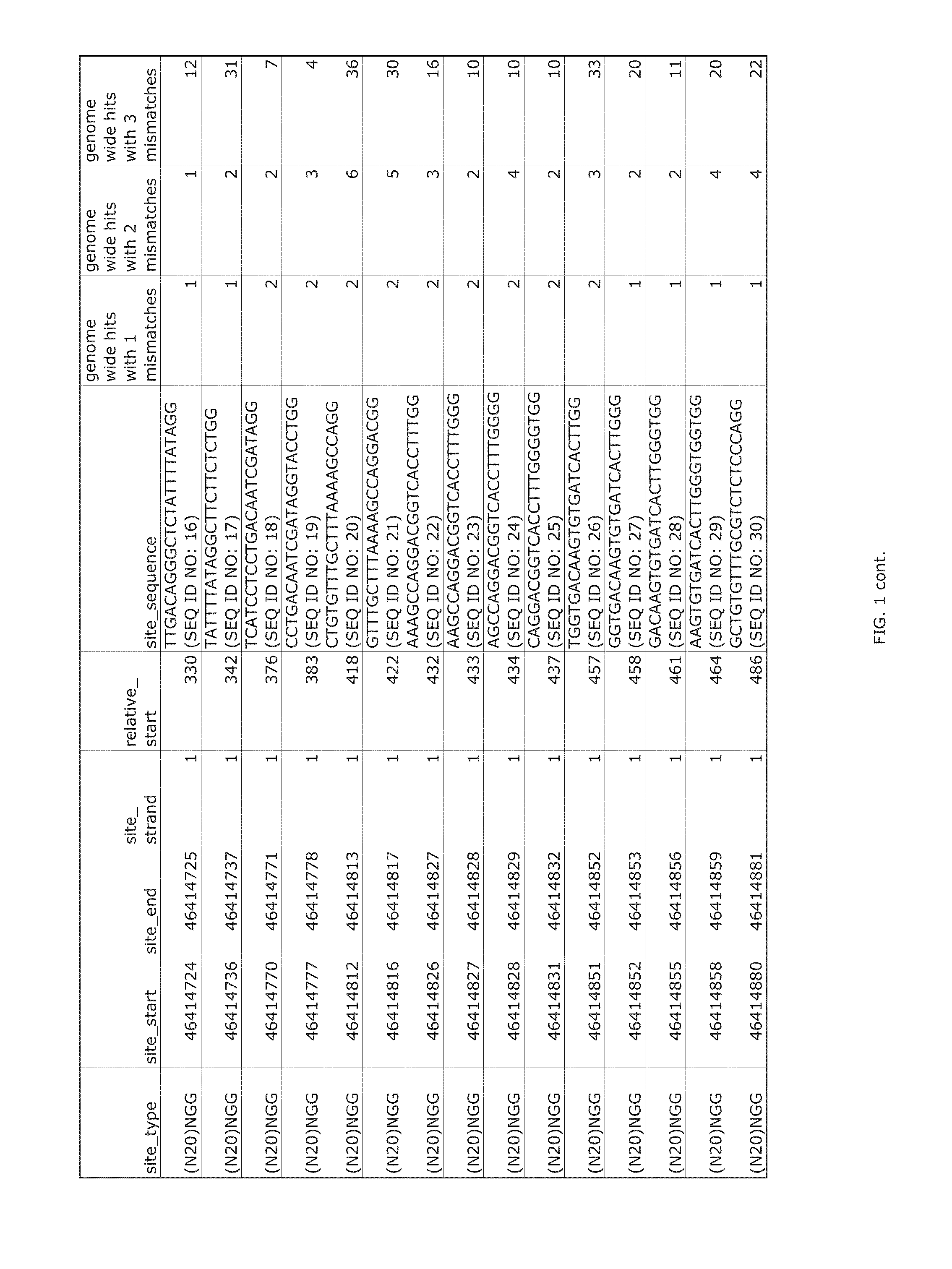
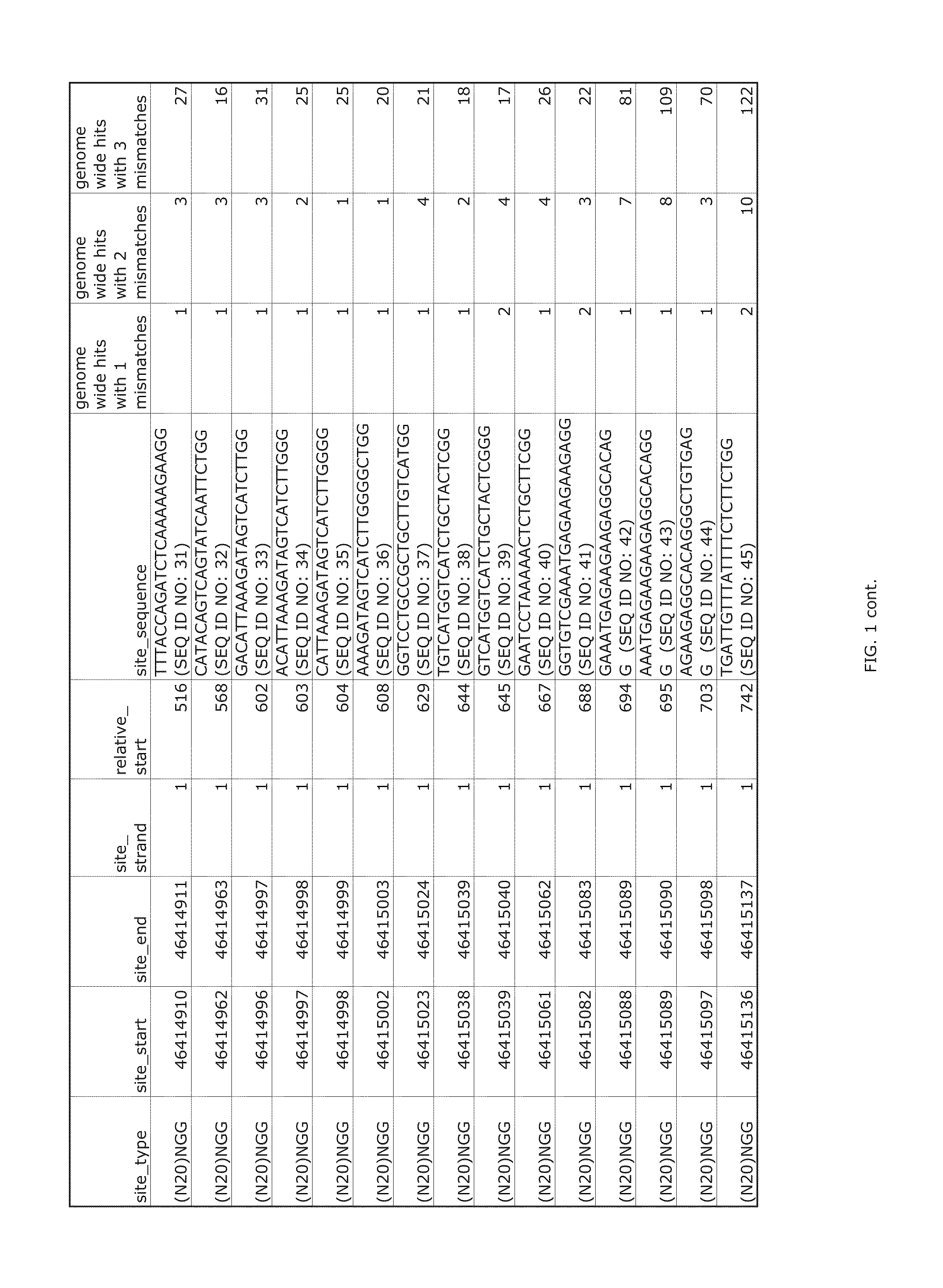
[0322]Transcription activator-like effector nucleases (TALENs) bind as a pair around a genomic site, in which a double-strand break (DSB) is introduced by a dimer of FokI nuclease domains. The use of a TALEN genome-editing system to rapidly and efficiently generate mutant alleles of 15 different genes in human pluripotent stem cells (hPSCs) as a means of performing rigorous disease modeling was recently reported (Ding et al., Cell Stem Cell 12:238-251 (2013)); the proportions of clones bearing at least one mutant allele ranged from 2%-34%.
[0323]As described below, the relative efficacies of CRISPRs and TALENs targeting the same genomic sites in the same hPSC lines was assessed with the use of the same delivery platform described previously (Ding et al., Cell Stem Cell 12:238-251 (2013)). In the TALEN genome-editing system, the CAG promoter was used to co-translate (via a viral 2A peptide) each TALEN with green fluorescent protein (GFP) or red fluorescent protein (RFP). For CRISPRs, ...
example 2
Efficient Targeting of Clinically Relevant Genes in Primary Somatic Cells
[0329]Work described herein shows for the first time that the CRISPR / Cas9 system can be used to edit the genome of somatic cells (e.g., primary) with high efficiency by using a double guide strategy. The inventors posit that this work will help bring genome editing in clinically relevant primary cells into reality.
[0330]The advent of genome editing tools that allow one to target any desired genomic site has greatly advanced the investigation of human biology and disease. In particular, the CRISPR / Cas9 system has become the gold standard in targeted genome editing technology, due to its flexibility and high efficacy. This system is constituted by the Cas9 nuclease from the microbial type II CRISPR / Cas system, which is targeted to specific genomic loci by a 20-nucleotide region in a synthetic guide RNA molecule. Similar to other targeted nucleases (ZFNs and TALENs), Cas9 induces double strand breaks (DSBs) that a...
example 3
Modified Cas9 mRNA Functions to Efficiently Introduce On-Target Mutations
[0356]The inventors generated Figment (Fgm) knockout mice by CRIPSR / Cas9 gene editing utilizing a modified Cas9 mRNA. Fgm is a coding gene within the long non-coding RNA Lnc-Rap-5 (referred to herein as Fgm (Lnc-Rap-5; see Sun et al., “Long noncoding RNAs regulate adipogenesis,” PNAS; 2013; 110(9):3387-3392, incorporated herein by reference in its entirety). The guide RNA (gRNA) sequence employed in this example was: 5′ gaggegaaagccactagcac 3′ (SEQ ID NO: 599). The modified Cas9 mRNA used in this example was made using an in vitro transcription reaction in which pseudouridine and 5-methyl-cytosine are reacted with unmodified nucleotides and randomly integrated into the resulting modified Cas9 mRNA. An exemplary protocol for generating Fgm knockout mice using CRISPR / Cas9 gene editing utilizing a modified Cas9mRNA is shown in FIG. 11A. As shown in FIG. 11A, 100 ng / μl of the resulting modified Cas9 mRNA and 50 ng / ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com