Method of increasing content of resistant starch in rice through genome editing and sgRNA special for same
A technology of resistant starch and gene editing, applied in the direction of recombinant DNA technology, DNA/RNA fragments, genetic engineering, etc., can solve the problems of non-improved agronomic traits, to reduce blood sugar, prevent hyperglycemia, prevent hyperglycemia and obesity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
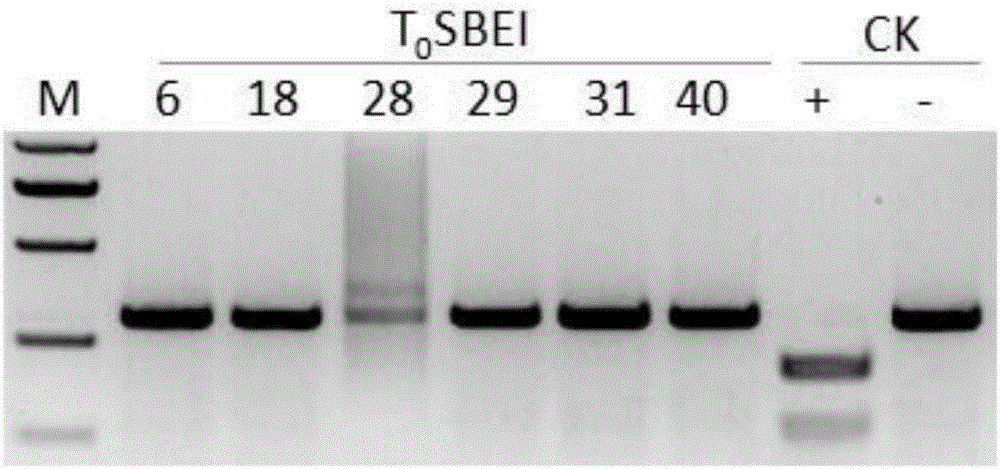
[0034] Embodiment 1, preparation recombinant plasmid
[0035] Artificially synthesized recombinant plasmid pCXUN-Cas9-gRNA1 (circular plasmid). The recombinant plasmid pCXUN-Cas9-gRNA1 is shown in Sequence 1 of the sequence listing. In sequence 1 of the sequence listing, the 109-361 nucleotides are reverse complementary to the NOS terminator, the 386-4516 nucleotides are reverse complementary to the coding gene of the Cas9 protein, and the 4537-6527 nucleotides are reverse complementary to the Cas9 protein encoding gene. The Ubi promoter is reverse complementary, nucleotides 6751-7154 are the U3 promoter, nucleotides 7155-7257 are the coding gene of sgRNA1 (the nucleotides 7155-7174 are the target sequence recognition region). The recombinant plasmid pCXUN-Cas9-gRNA1 expressed sgRNA1, and the target sequence of sgRNA1 was located in the first exon of rice SBEI gene. The rice SBEI gene is shown in sequence 3 of the sequence table (genomic DNA), the 1875-1958th nucleotide in t...
Embodiment 2
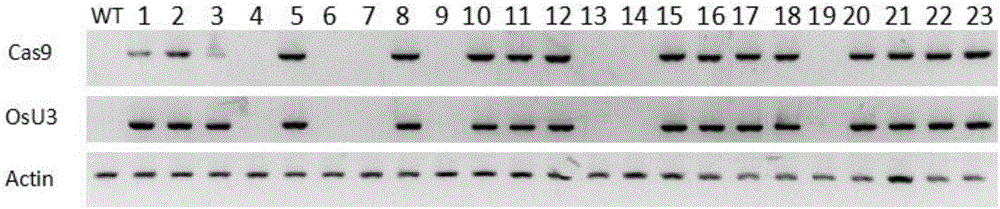
[0037] Embodiment 2, use recombinant plasmid to prepare transgenic plant
[0038] 1. Obtaining of transgenic plants
[0039] 1. Introduce the recombinant plasmid pCXUN-Cas9-gRNA1 into Agrobacterium EHA105 to obtain recombinant Agrobacterium.
[0040] 2. Using rice kitaake as the starting plant, using the recombinant Agrobacterium obtained in step 1, performing Agrobacterium-mediated genetic transformation to obtain T 0 generation of regenerated plants.
[0041] The specific steps of genetic transformation are as follows:
[0042] (1) Resuspend the recombinant Agrobacterium obtained in step 1 with AAM medium to obtain OD 600nm =0.3-0.5 bacterial suspension.
[0043] AAM medium (pH5.2): 4.3g / L MS salts&vitamins+68.5g / L sucrose+0.5g / LMES+36g / L glucose+500mg / L casamino acids+100ml 10×AA amino acids solution+40mg / L acetosyringone. 10×AA amino acids solution: 8.76g / L L-glutamine, 2.66g / L L-aspartic acid, 1.74g / LL-arginine and 75mg / L glycine.
[0044] (2) Take rice kitaake se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com