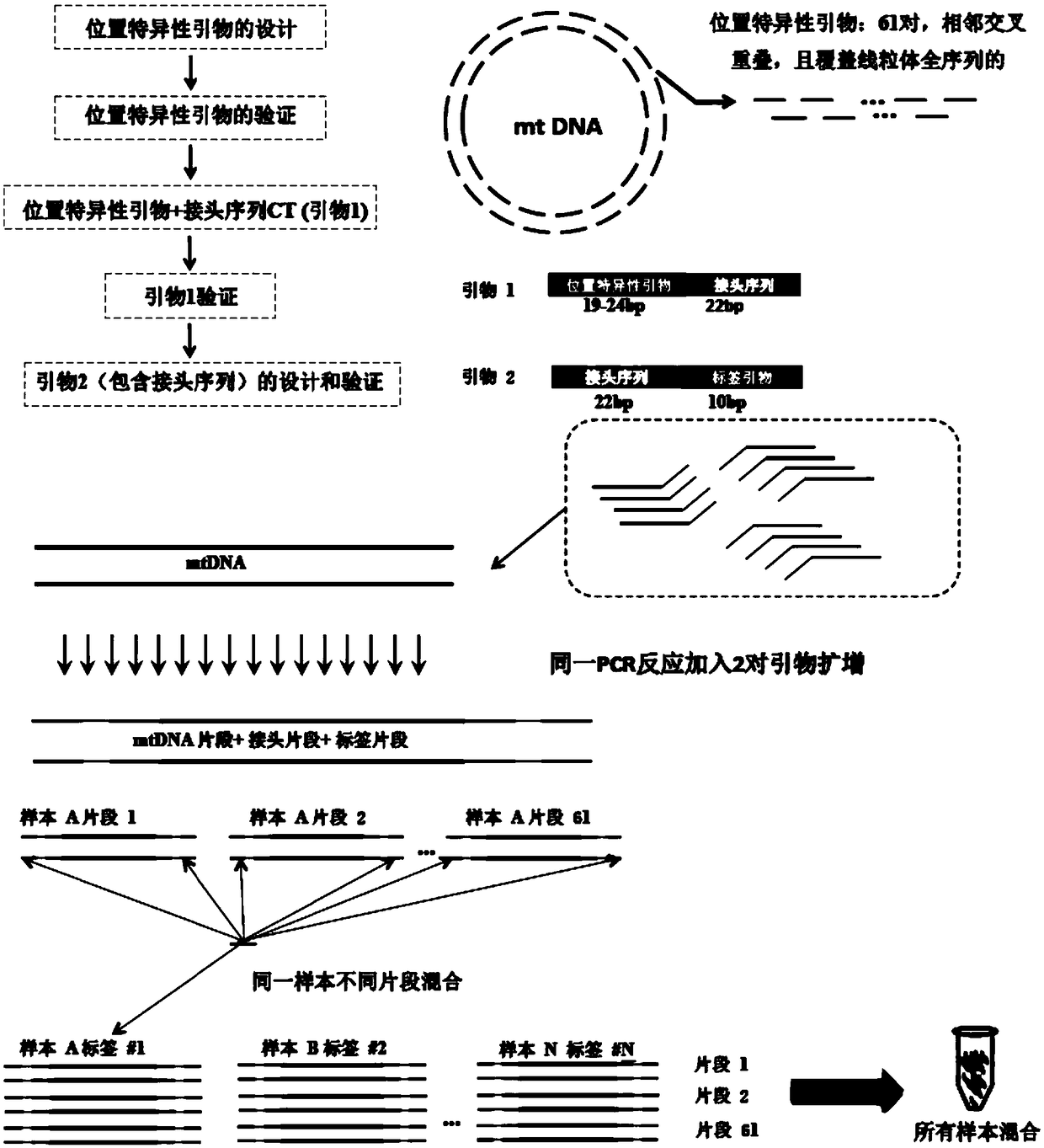
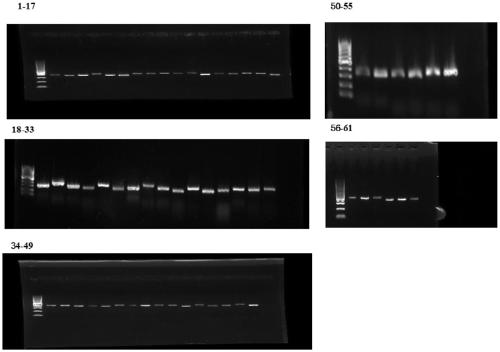
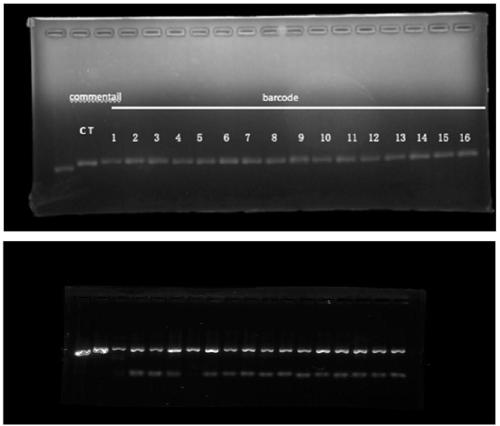
Mitochondria whole genome sequencing method based on high-throughput sequencing
A whole genome and mitochondrial technology, applied in the field of genetic engineering, can solve the problems of increased workload, primer cost and barcode sequence consumption, etc., to achieve the effect of reducing workload, reducing sequencing cost and time, and increasing the number
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0033] The technical solution of this aspect will be further described in detail below in conjunction with the accompanying drawings and specific embodiments, but the present invention is not limited to the following embodiments. Get 16 samples, carry out the verification of the present invention, the specific steps of experimental process are as follows.
[0034] 1. Extraction and preservation of mtDNA template
[0035] 1.1 Sample collection and storage.
[0036] Peripheral venous blood was collected from each subject, anticoagulated with EDTA, and stored in a 4°C refrigerator.
[0037] 1.2 DNA extraction
[0038] Follow the routine whole blood DNA extraction procedure.
[0039] 1.3 DNA sample quality control
[0040] Take 1 ul of DNA sample, and test the concentration and purity of DNxA template by spectrophotometry, the concentration is ≥20ng / ul, 260 / 280 is 1.8-2.0; 260 / 230 2.0-2.5 samples are considered qualified. Quality control qualified samples were stored at -20°C...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com