System and Methods for Detecting Genetic Variation
a genetic variation and system technology, applied in the field of system and methods for detecting genetic variation, can solve the problems of reducing reliability and reproducibility, reducing the efficiency of the process, and a large amount of genomic sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Sample Preparation and Sequencing Process
[0167]Genomic DNA (gDNA) is extracted in 96-well format, leaving wells A1, G12, and H12 empty (which will later contain a no-template control, the universal negative standard containing Coriell sample NA12878 genomic DNA lacking every causal genetic variant tested, and a sample comprising one of a plurality of known causal genetic variants, respectively). 50 μL from each well are transferred into a corresponding well of an absorbance plate. Absorbance at 260 nm is measured using a Tecan M200 plate reader to calculate DNA quantity. 50 μL of gDNA are transferred from the absorbance plate into an Eppendorf twin.tec plate. Control samples are added to their respective position on the twin.tec plate. The gDNA and controls are fragmented in a SonicMan (Matrical, Spokane Wash.) sonicator, according to the following protocol at 10° C.: Pre-chill 180 s, cycles 100, sonication 3.0s, power 35%, lid chill 1.0 s, plate chill 0, post chill 0. A 2 μL sample...
example 2
Amplification and Sequencing Process
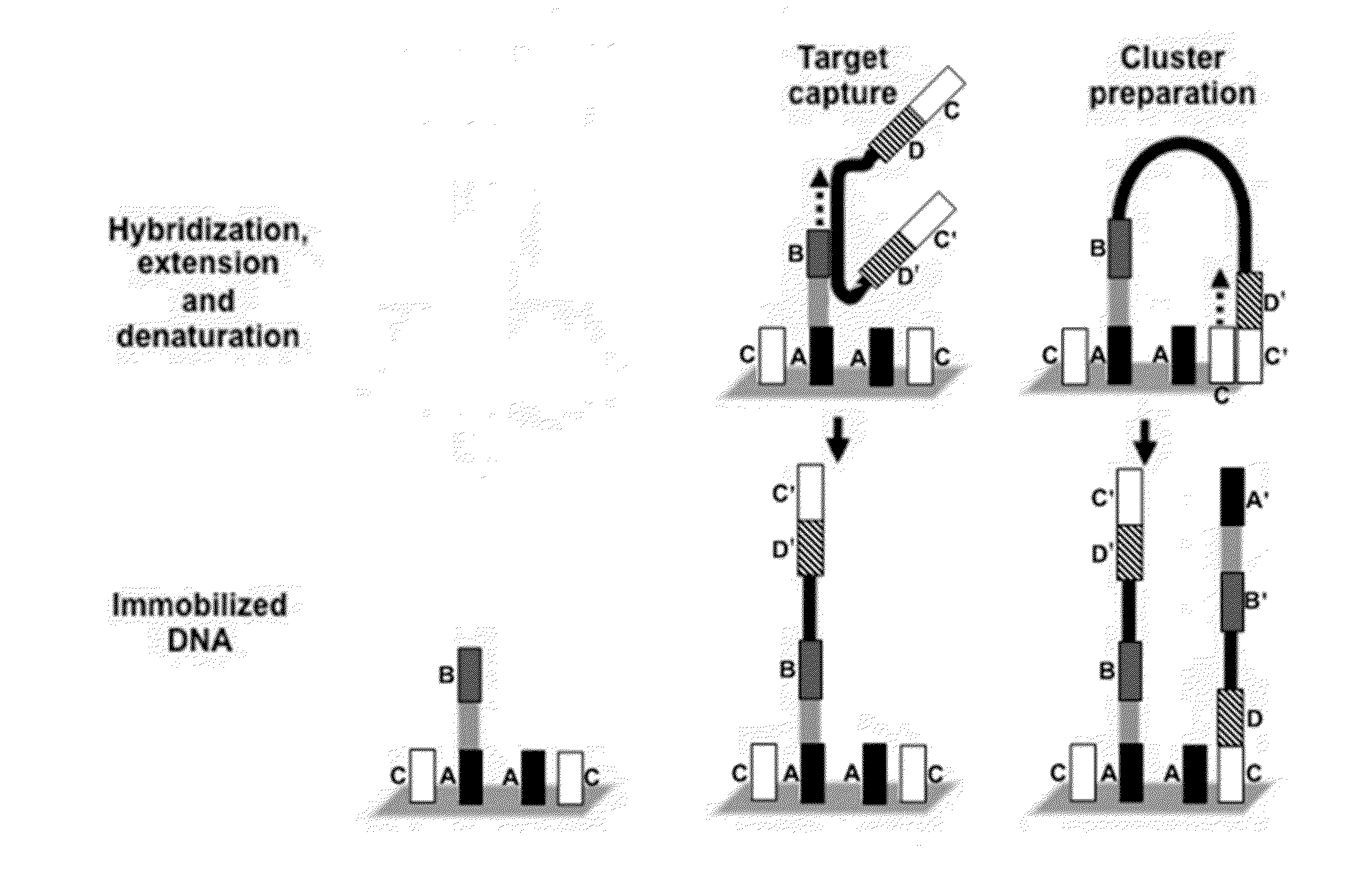
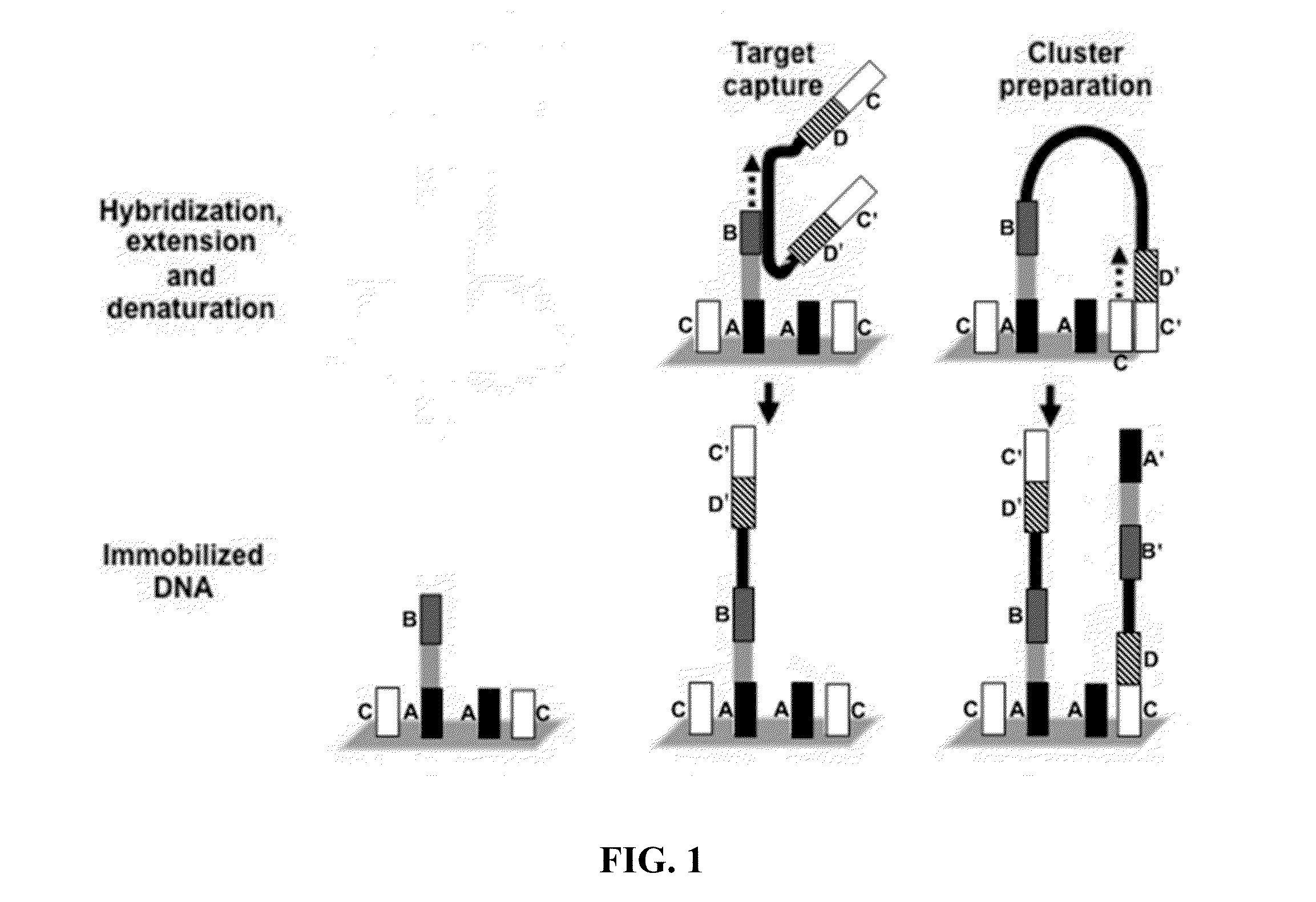
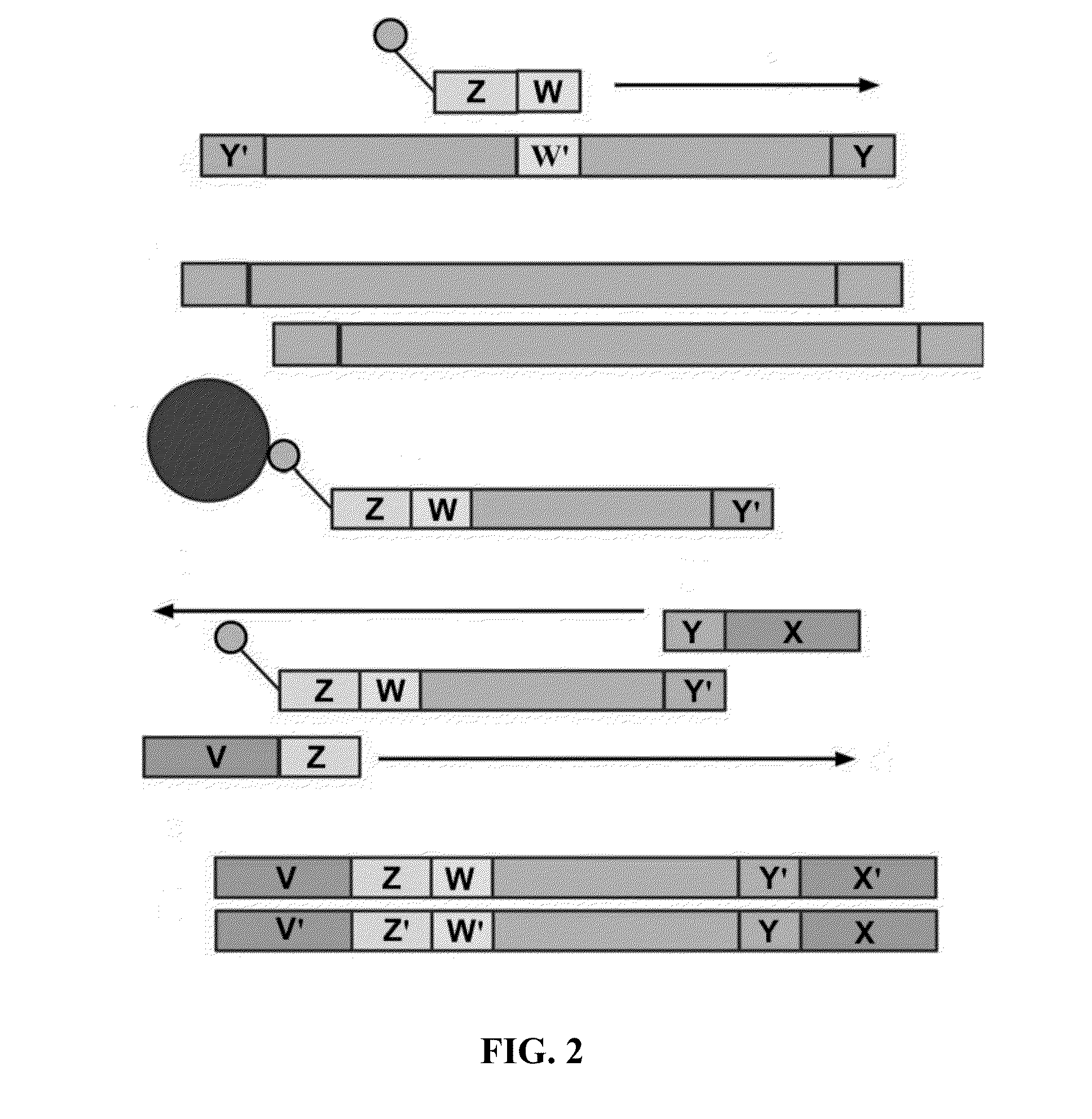
[0174]Example processes for the amplification of a plurality of different target polynucleotides are illustrated in FIGS. 2 and 5, which differ primarily in the inclusion of a solid-phase purification step in FIG. 2. FIG. 7 also illustrates an example amplification process, and differs from the process illustrated in FIG. 2 primarily in that oligonucleotide primer extension is performed before adapter joining, instead of after adapter joining. Amplification may or may not include a solid-phase purification step. FIG. 6 illustrates an amplification process as in FIG. 5, and also example bridge amplification and sequencing processes. The amplification process illustrated in FIG. 6 may be used in conjunction with any bridge amplification method and associated sequencing method.
[0175]First, a partially single-stranded adapter is ligated to fragmented polynucleotides. The partially single-stranded adapter has a double-stranded region at one end (sequen...
example 3
Identification of Non-Subject Sequences
[0177]Polynucleotides (e.g. DNA and / or RNA) are extracted from a sample from a subject suspected to contain viral and / or bacterial polynucleotides using standard methods known in the art. Sample polynucleotides are fragmented, end-repaired, and A-tailed, such as in Example 1. Adapter oligonucleotides comprising sequence D are then joined to the sample polynucleotides, which are then amplified using amplification primers comprising sequence C, sequence D, and a barcode. Amplified target polynucleotides are hybridized to a plurality of different first oligonucleotides that are attached to a solid surface. Each first oligonucleotide comprises sequence A and sequence B, where sequence B is different for each different first oligonucleotide, is at the 3′ end of each first oligonucleotide, and is complementary to a sequence comprising a non-subject sequence or a sequence within 200 nucleotides of a non-subject sequence. Specifically, the first oligon...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com