Primer group and method for rapidly identifying influenza A virus subtypes through combination of DNA barcodes and second-generation high-throughput sequencing
An influenza virus, high-throughput technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems that the development of barcode technology has not yet been reported, and achieve the goal of rapid influenza typing diagnostic procedures Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
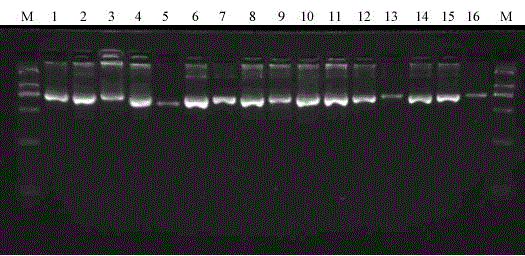
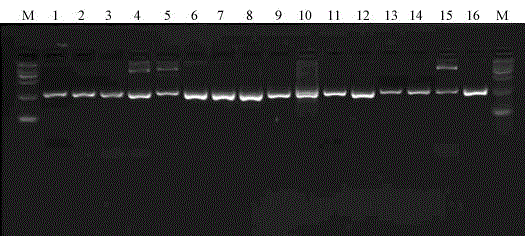
[0049] This example provides a method for rapidly identifying influenza A virus subtypes by using DNA barcodes combined with next-generation high-throughput sequencing.
[0050] 1 Materials and methods
[0051] 1.1 Strain materials (09pdm swine flu H1N1, equine flu H3N8, bird flu H2N9, H4N1, H5N1, H7N2, H9N2, H13N6, H16N3) inactivated samples are kept by our laboratory, H6N5, H8N4, H10N8, H11N6, H12N5, H14N5, The H15N8 nucleic acid sample was donated by the Jilin Entry-Exit Quarantine Bureau (see the table below for details)
[0052] No.
Isolate
Acc. no.
1
A / swine / Shandong / 01 / 2009(H1N1)
KM368313
2
A / duck / Nanchang / 4-190 / 2000(H2N9)
CY117211
3
A / equine / Haerbin / 01 / 2010(H3N8)
KM892526
4
A / duck / Hong Kong / 951 / 1980(H4N1)
AB292404
5
A / chicken / Shandong / K01 / 2004(H5N1)
DQ767725
6
A / duck / Guangxi / 1455 / 2004(H6N5)
HM144533
7
A / duck / Hong Kong / 293 / 78(H7N2)
CY006029
8 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com