Method for detecting mutation information in multiplex amplification sequencing product of genome
A technology for multiple amplification and detection methods, which is applied in the field of detection of mutation information in multiple genome amplification and sequencing products, and can solve the problems of slow program running, not considering differences in gene expression patterns, and being susceptible to interference.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
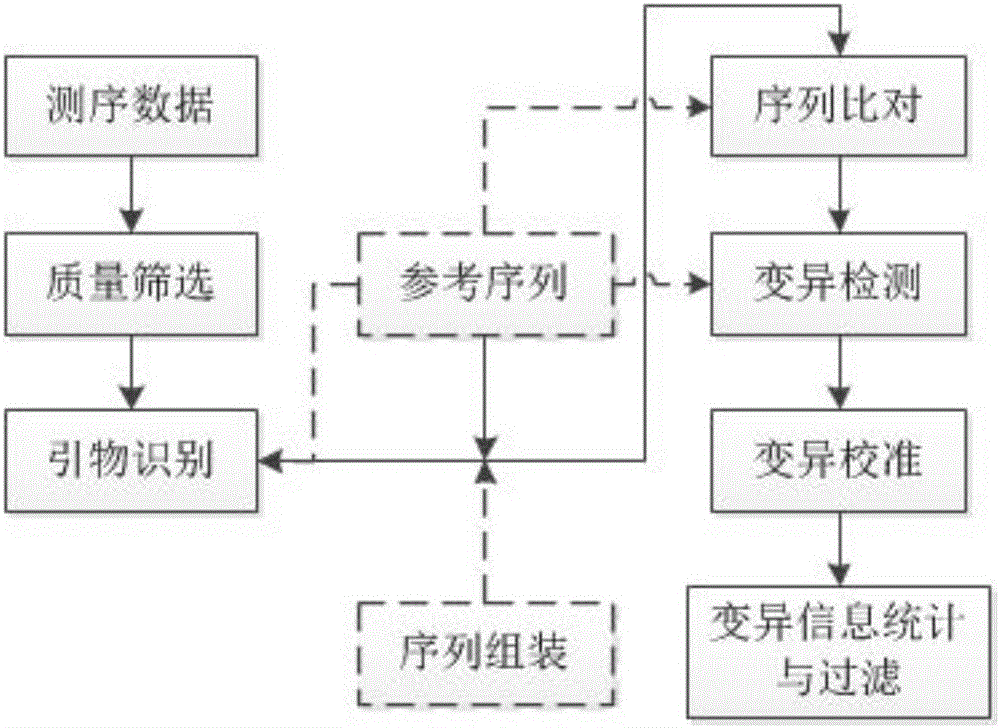
Image
Examples
Embodiment 1
[0167] Example 1: Method system for detecting mutation information in multiple amplification sequencing products of human genome
[0168] Using the method of the present invention, 107 cases of BRCA1 and BRCA2 gene sequencing data obtained by multiple amplification were analyzed. These included 100 whole blood samples from healthy voluntary blood donors, 5 breast or ovarian cancer patient samples (whole blood, paraffin section), 2 strains of BRCA1 / 2 positive cell lines, respectively BT474 and HCT15 (both available from ATCC).
[0169] (1) Quality assessment and preprocessing of sequencing data
[0170] The 107 PE250 data obtained by the illumina sequencing platform were subjected to low-quality screening and low base recognition screening. The specific filtering conditions are: Q20 is lower than 80% or the proportion of N bases is greater than 20%. Finally, all samples had high sequencing data quality, see Table 1 for details.
[0171] Table 1. Statistical results of basic i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com