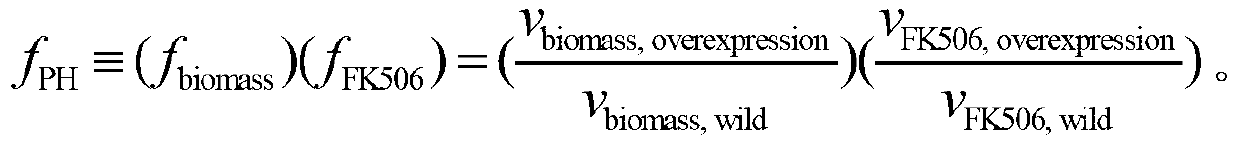Patents
Literature
52 results about "Genome comparison" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Comparing genomes (entire DNA sequences) of different species provides a powerful new tool to explore these relationships. Genomes are more than instruction books for building and maintaining an organism; they contain vast amounts of information on the history of life.
Gene expression profiling based identification of genomic signature of high-risk multiple myeloma and uses thereof
InactiveUS20080274911A1High-risk indexIncreased risk of deathMicrobiological testing/measurementLibrary screeningNewly diagnosedComparative genomic hybridization
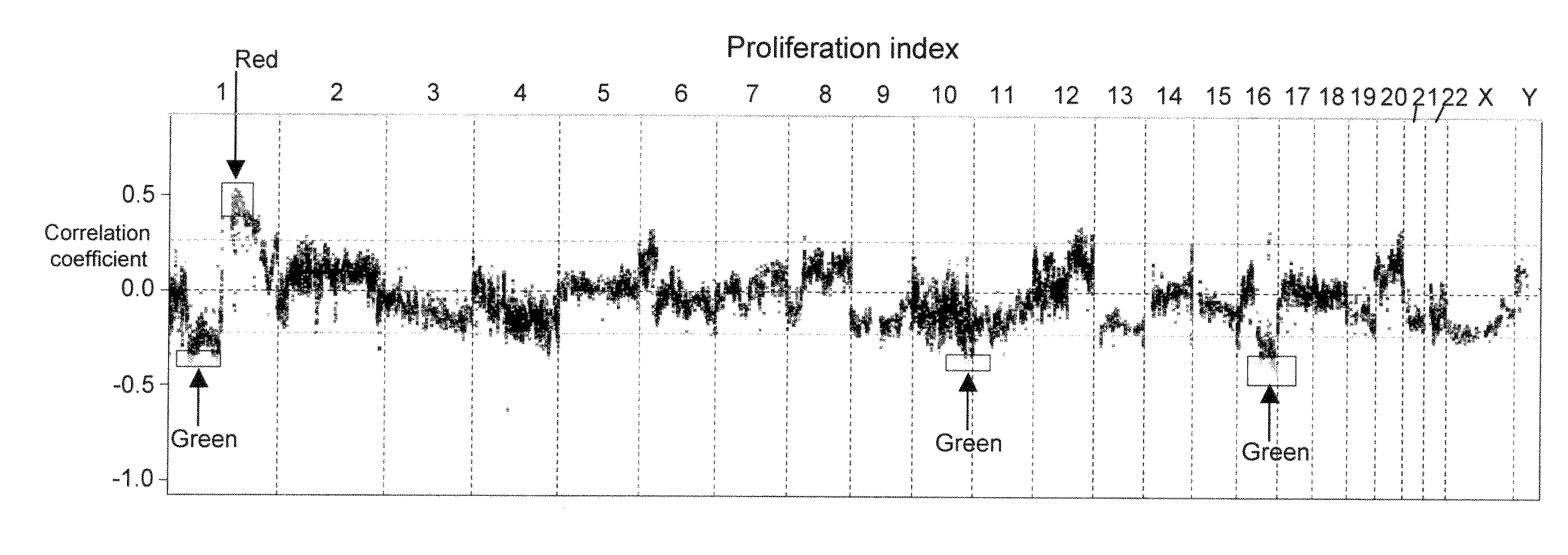
The present invention discloses a method of applying novel bioinformatics and computational methodologies to data generated by high-resolution genome-wide comparative genomic hybridization and gene expression profiling on CD138-sorted plasma cells from a cohort of 92 newly diagnosed multiple myeloma patients treated with high dose chemotherapy and stem cell rescue. The results revealed that gains the q arm and loss of the p arm of chromosome 1 were highly correlated with altered expression of resident genes in this chromosome, with these changes strongly correlated with 1) risk of death from disease progression, 2) a gene expression based proliferation index, and 3) a recently described gene expression-based high-risk index. Importantly, we also found a strong correlation between copy number gains of 8q24, and increased expression of Argonate 2 (AGO2) a gene coding for a master regulator of microRNA expression and maturation, also being significantly correlated with outcome. Our novel findings significantly improve our understanding of the genomic structure of multiple myeloma and its relationship to clinical outcome.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ARKANSAS
Cell subset annotation method based on single cell transcriptome sequencing
ActiveCN112700820AComprehensive identificationFix the annotation problemBiostatisticsSequence analysisSingle cell transcriptomeCell subpopulations
The invention provides a cell subset annotation method based on single cell transcriptome sequencing. The cell subset annotation method comprises the following steps: 1) 10x barcode UMI identification; 2) genome comparison; 3) gene expression profile comparison; 4) low-quality cell filtering and data homogenization; 5) cell population clustering; 6) marker gene extraction; 7) cell subset annotation. The invention belongs to the technical field of biological information analysis, and provides a cell subset annotation method based on single-cell transcriptome sequencing, which solves the problem of single-cell subset annotation, so that single-cell sequencing data can support cell annotation according to a gene expression profile and / or a cell Marker gene after conventional analysis. Therefore, organic combination of different annotation methods is realized, and the distribution condition and related information of cell types are obtained.
Owner:广州华银医学检验中心有限公司 +1
Gene chip for predicting liver cancer metastasis and recurrence risk and manufacturing method and using method thereof
InactiveCN101812507AImprove survivalAccurate predictionMicrobiological testing/measurementLymphatic SpreadCvd risk
The technical scheme of the invention provides a gene chip for predicting liver cancer metastasis and recurrence risk, which comprises a substrate and a probe arranged on the substrate, wherein the probe consists of 149 genes. The gene chip for predicting the liver cancer metastasis and the recurrence risk has the advantages that: due to the adoption of genome comparison research, a large sample independently proves that a prediction model can accurately predict and evaluate the metastasis potential of liver cancer patients. Therefore, postoperative survival and metastasis, and recurrence of the patient can be accurately predicted (even early liver cancer patients can be accurately predicted). The gene chip contributes to early identifying or predicting high-risk patients so as to perform intensive monitoring and effective intervention on the high-risk patients to further prolong the life of the tumor patients.
Owner:ZHONGSHAN HOSPITAL FUDAN UNIV
Specific DNA fragment SSM2 for sex determination of sturgeons and application
ActiveCN111471775AReduce harmMicrobiological testing/measurementDNA/RNA fragmentationZooidAnimal science
The invention belongs to the field of fish sex determination in the field of aquaculture, and particularly relates to a specific DNA fragment SSM2 for sex determination of sturgeons and application. According to the invention, a specific DNA fragment of sturgeon female individuals is obtained through a high-throughput sequencing and genome comparison method. Primers are designed for the fragment,and can perform accurate and rapid sex identification on nine kinds of sturgeons. An identification method of the invention has the characteristics of simple, quick and accurate operation, extremely small damage to the sturgeons and the like, solves the problem of sturgeon sex identification, and is helpful for rapid development of the sturgeon culture industry.
Owner:YANGTZE RIVER FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
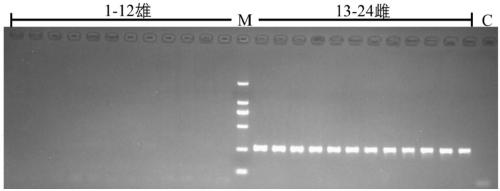
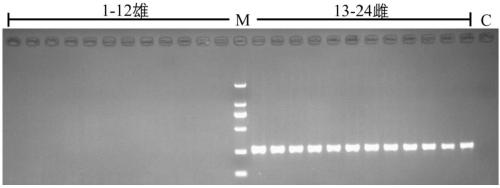
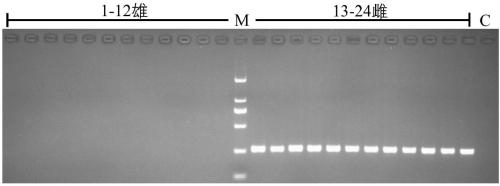
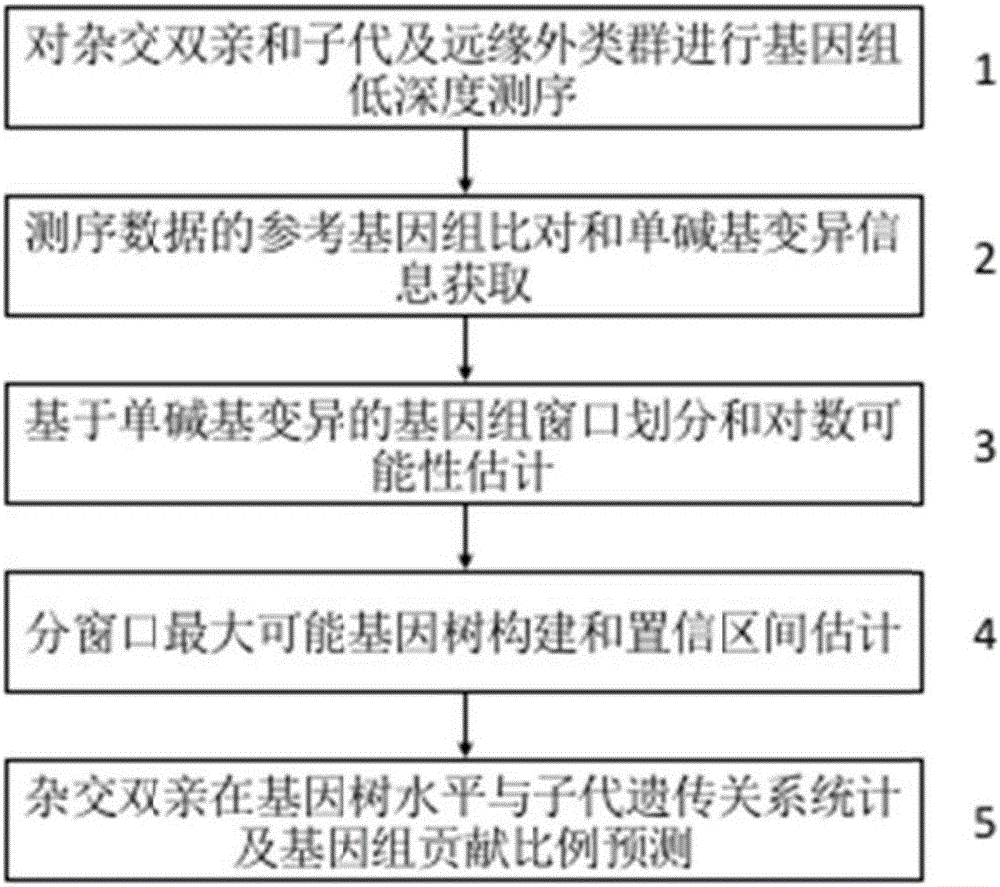
Method for recognizing contribution proportion of kiwi fruit hybrid patients on filial generation genome
ActiveCN106755300AImprove accuracyFacilitate early screeningMicrobiological testing/measurementHybridisationConfidence intervalKiwi fruit
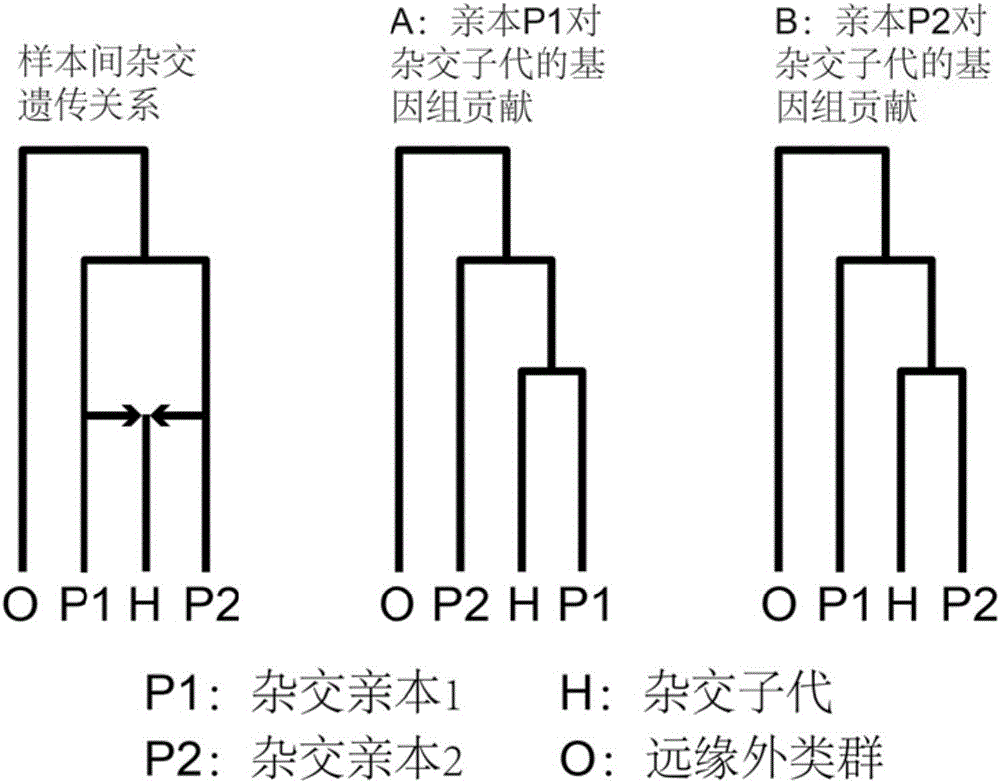
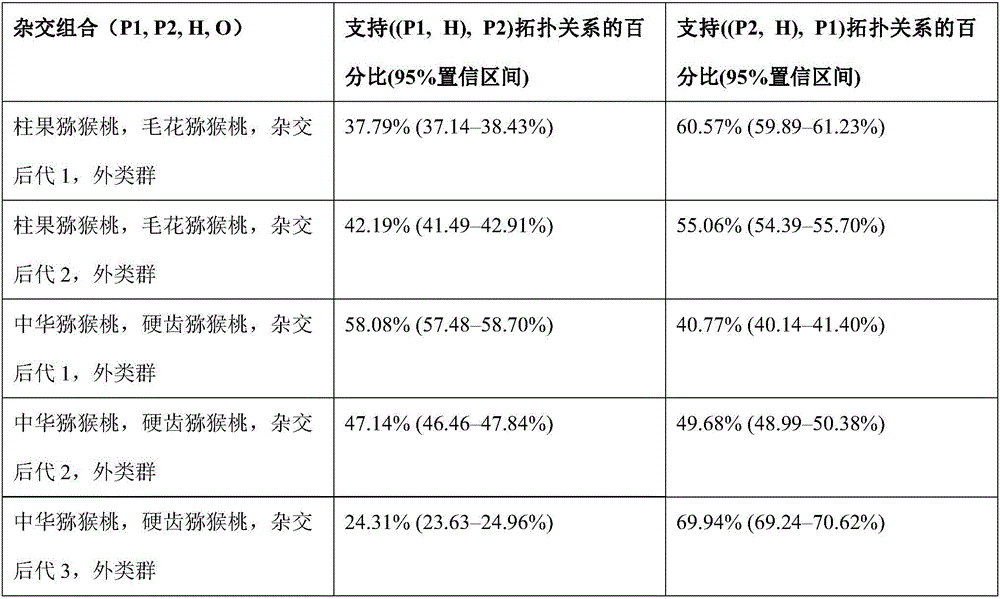
The invention discloses a method for recognizing the contribution proportion of kiwi fruit hybrid patients on a filial generation genome. The method comprises the following steps of performing genome low-depth sequence testing on hybrid parents and filial generations and distant hybrid outgroup; performing sequence testing data reference genome comparison, and obtaining single basic group variation information; performing single basic group variation-based genome window division and log probability estimation; performing sub window maximum possible gene tree building and confidence interval estimation; performing gene tree level and filial generation genetic relationship statistics and genome contribution ratio prediction on the hybrid parents. The whole genome variation information is used for analyzing the evolution genetic relationship of the hybrid patients and the hybrid filial generation, so that the prediction of the hybrid filial generation characteristic properties realizes high accuracy; meanwhile, by using the method, the magnitude and the direction of the possibly existing phenotypic characteristic variation are predicted in the baby period of the hybrid filial generation formation; the early stage screening of the hybrid strains can be greatly promoted; the labor and material cost can be reduced; the resource mining and utilization efficiency can be greatly improved.
Owner:SOUTH CHINA BOTANICAL GARDEN CHINESE ACADEMY OF SCI
Specific primer for novel coronary virus detection and rapid detection method
ActiveCN111349719AAids in determining infectionAccurate detectionMicrobiological testing/measurementAgainst vector-borne diseasesVirus detectionBioinformatics
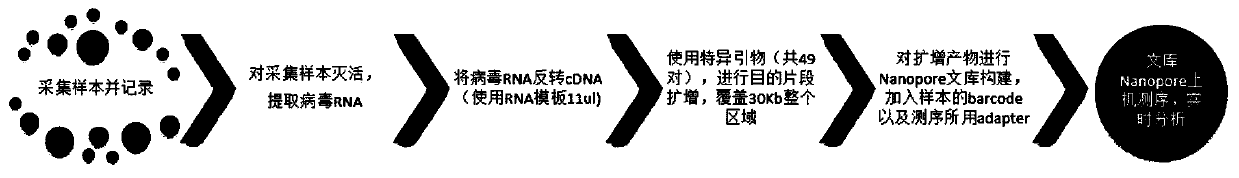
The invention discloses a specific primer for novel coronary virus detection and a rapid detection method. The method comprises the following steps of inactivating a collected sample, and extracting virus RNA; carrying out inversion on virus RNA which is extracted in the step S1 and qualified through quality testing to perform cDNA synthesis; adopting the specific primer for fragment amplificationto form a PCR reaction product sample; identifying the PCR reaction product sample, and constructing a sequencing library; and carrying out a nanopore sequencing operation on the sequencing library.The method is high in sequencing quality, and when the sample obtained by the method is applied to novel coronary virus genome comparison, reads comparison rate as high as 94.16% can be realized, so that determination of the infection condition of a patient can be assisted, and rapid accurate detection of novel coronary virus can be realized.
Owner:THE FIRST AFFILIATED HOSPITAL OF GUANGZHOU MEDICAL UNIV (GUANGZHOU RESPIRATORY CENT) +2
Specific DNA fragment SSM1 for sex determination of sturgeons and application
ActiveCN111471756AReduce harmMicrobiological testing/measurementDNA/RNA fragmentationRapid identificationSturgeon
The invention belongs to the field of fish sex determination in the field of aquaculture, and particularly relates to a specific DNA fragment SSM1 for sex determination of sturgeons and application. Asturgeon female sex specific DNA fragment SSM1 is successfully screened through high-throughput sequencing and genome comparison. Primers are designed and can accurately and rapidly identify the sexes of nine kinds of sturgeons. The invention also discloses primer sequences, and PCR reaction system and conditions. An identification method of the invention has the characteristics of simple, quickand accurate operation, extremely small damage to the sturgeons and the like, solves the problem of sturgeon sex identification, and is helpful for rapid development of the sturgeon culture industry.
Owner:YANGTZE RIVER FISHERIES RES INST CHINESE ACAD OF FISHERY SCI
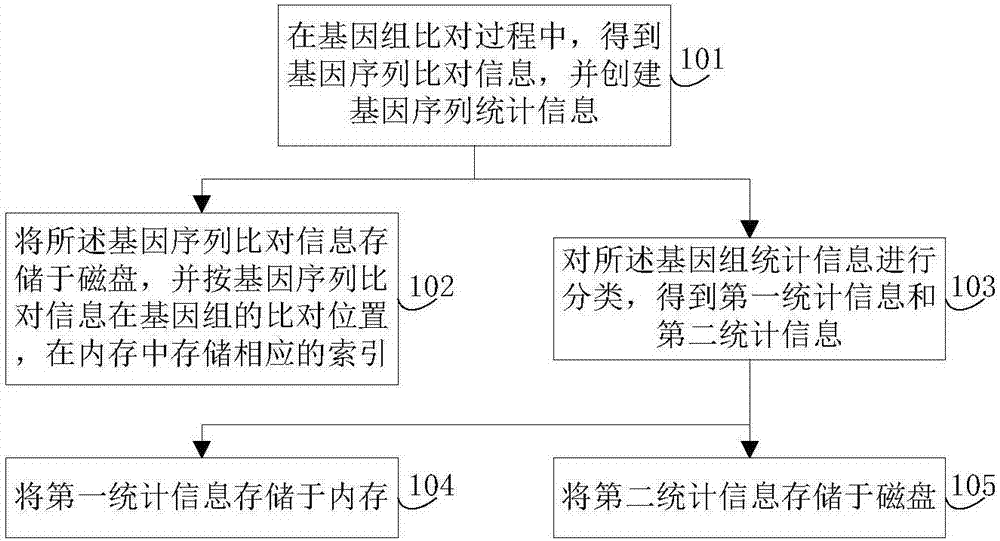
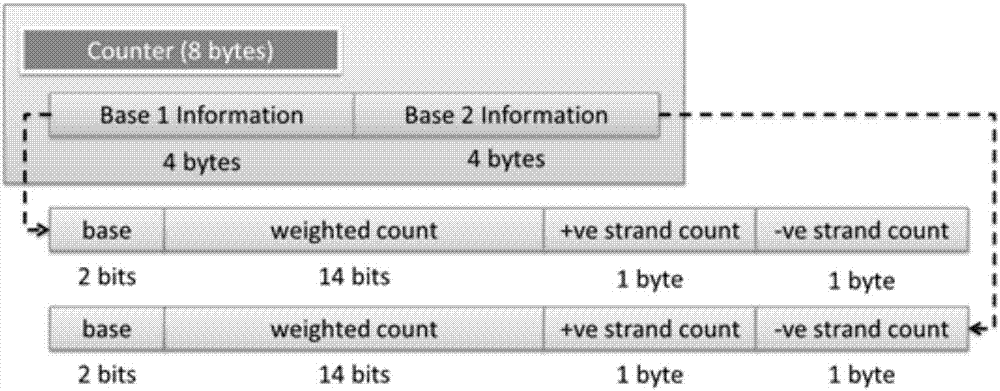
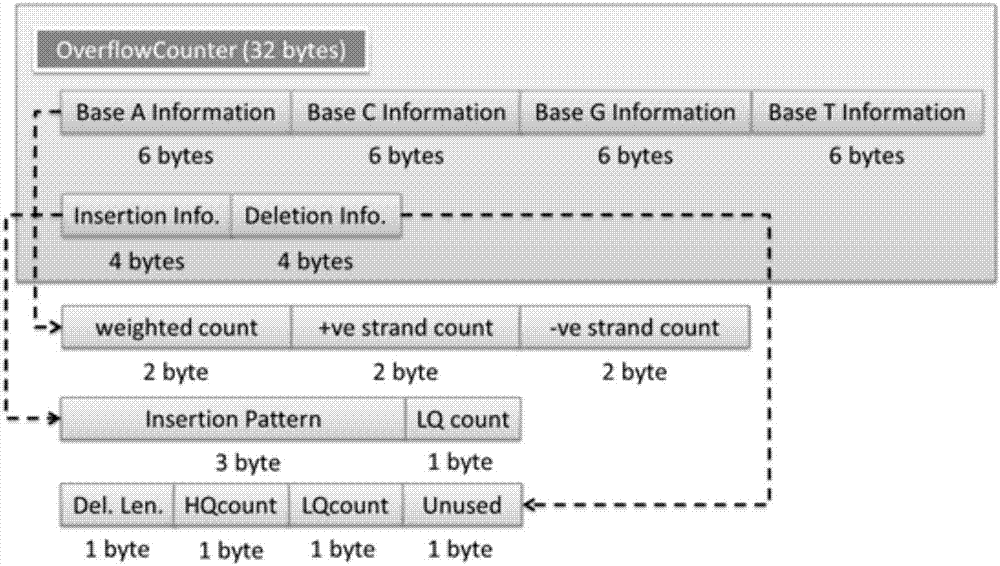
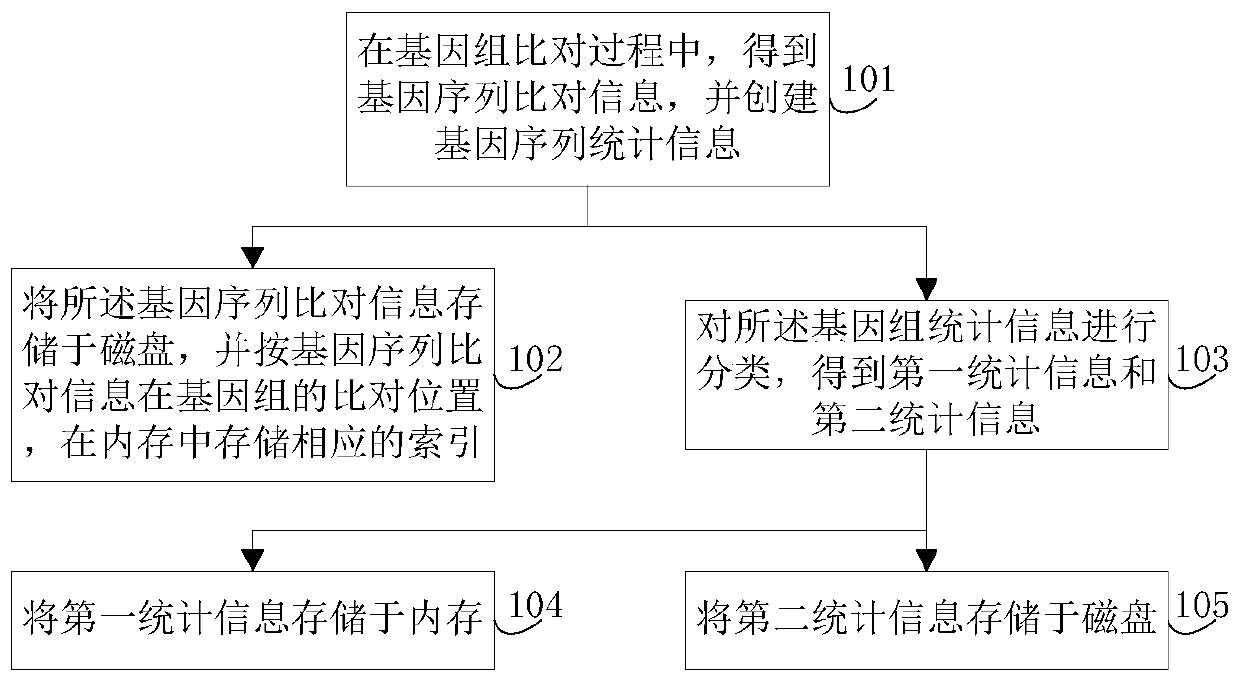
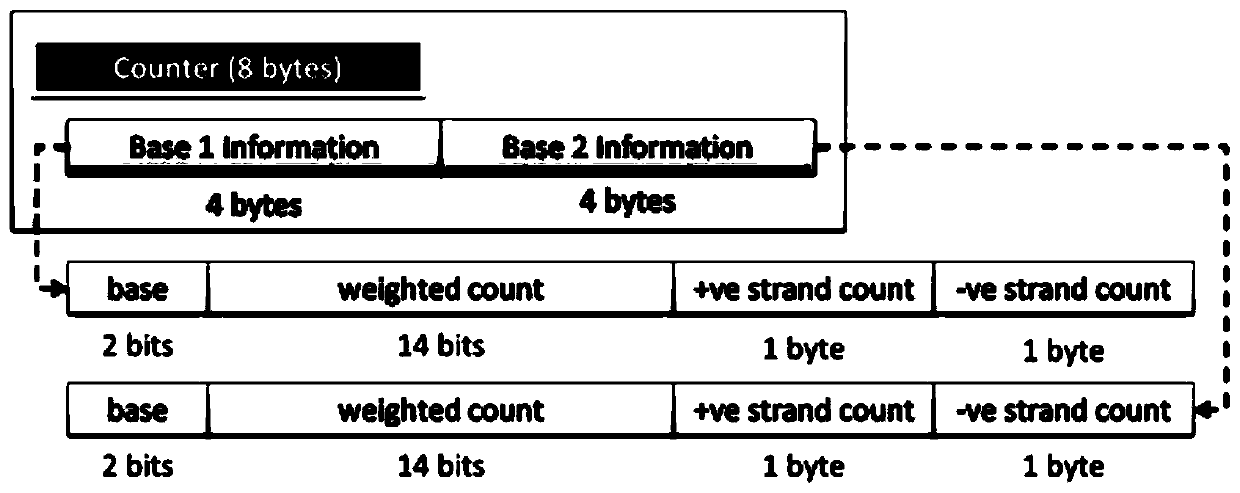
Genomic-data storage method and electronic device
ActiveCN107480466AImprove efficiencyInput/output to record carriersSpecial data processing applicationsGenome alignmentAccess frequency
The invention discloses a genomic-data storage method. The genomic-data storage method includes the steps that in the genome comparison process, gene-sequence comparison information is obtained, and gene-sequence statistical information is established; the gene-sequence comparison information is stored in a magnetic disk, and according to comparison positions of the gene-sequence comparison information in a genome, corresponding indexes are stored in a memory, wherein the indexes are storage positions of the gene-sequence comparison information in the magnetic disk; genome statistical information is classified, and first statistical information and second statistical information are obtained; the first statistical information is stored in the memory, wherein the first statistical information is statistical information with the access frequency higher than the preset frequency in the variation detection process; the second statistical information is stored in the magnetic disk, wherein the second statistical information is statistical information which cannot be stored in the memory and / or statistical information with the access frequency lower than the preset frequency in the variation detection process. The invention also discloses an electronic device with the genomic-data storage method.
Owner:UNITED ELECTRONICS
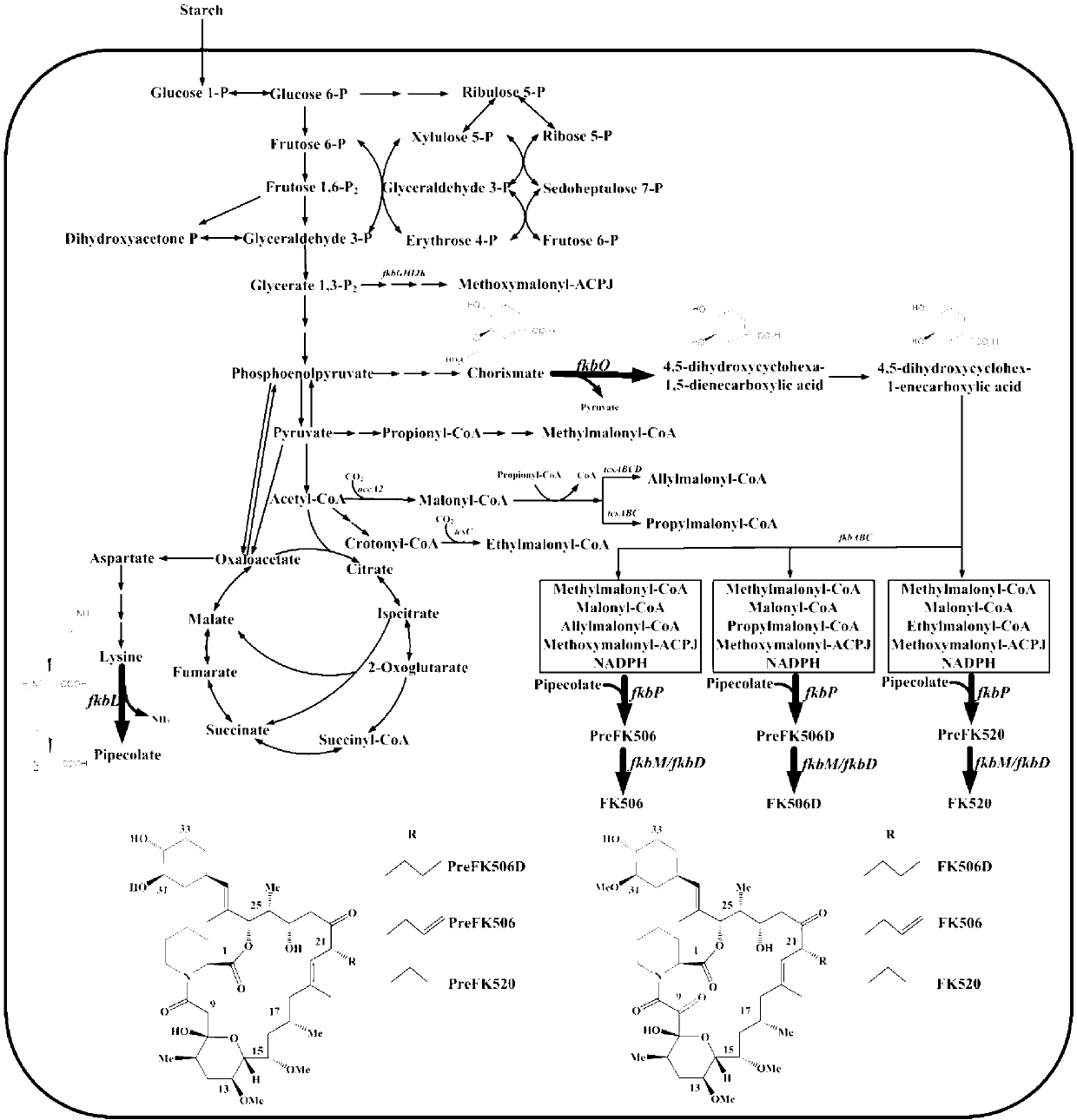
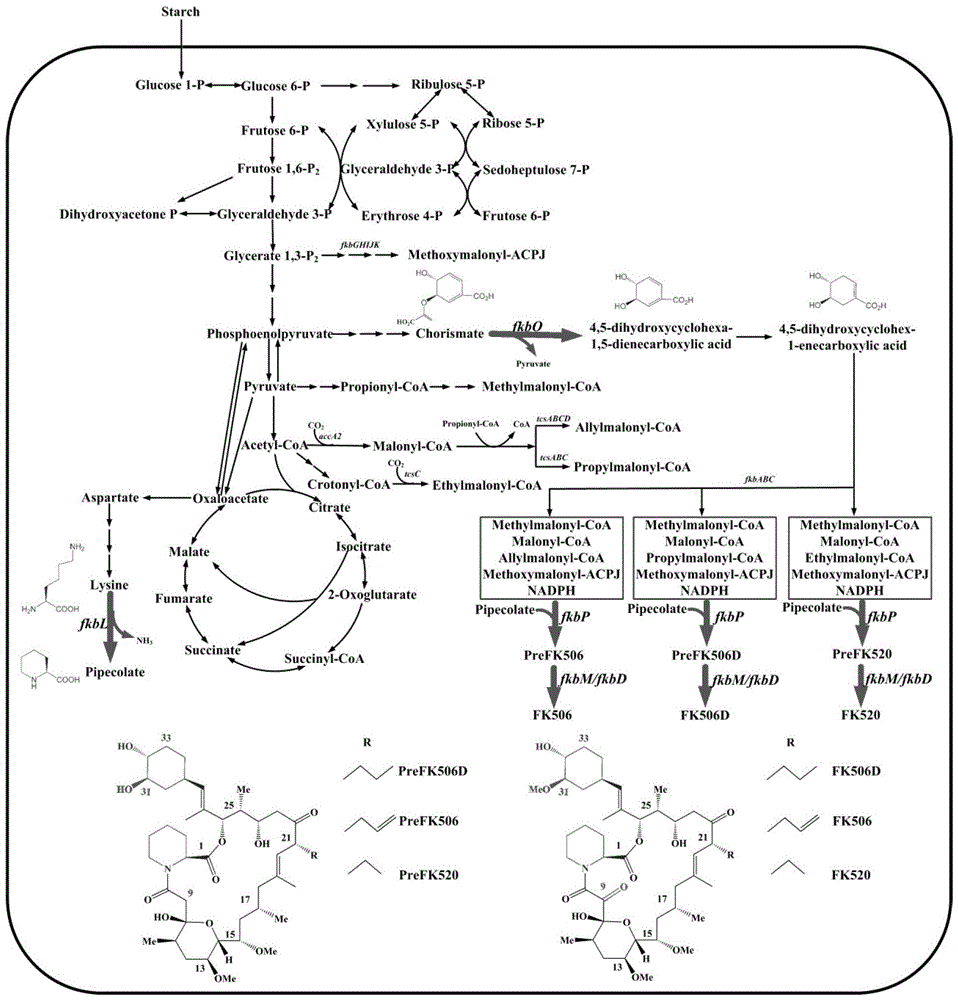
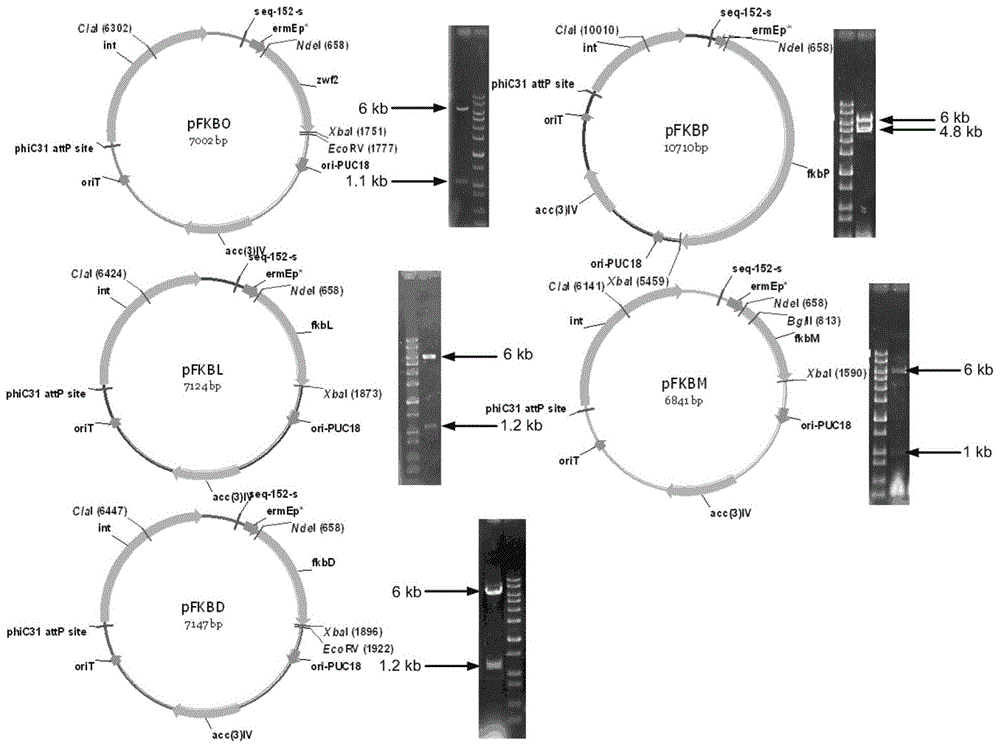
Secondary approach transformation method based on instruction of FK506 production bacterial strain wave chain streptomycete genome scale metabolic network model
InactiveCN103279689AIncrease production levelsImprove the experimental effectSpecial data processing applicationsMicroorganismGene cluster
The invention discloses a secondary approach transformation method based on an instruction of an FK506 production bacterial strain wave chain streptomycete genome scale metabolic network model. The model is based on annotation genes and physiology and biochemistry information. By comparing and analyzing the model with a streptomyces coelicolor genome, metabolic genes are found being highly conservative. Metabolic flux analysis is performed on a genome scale metabolic network, and therefore the model predicts a mutation bacterium secondary approach gene cluster transformation strategy for improving a production level. According to the secondary approach transformation method based on the instruction of the FK506 production bacterial strain wave chain streptomycete genome scale metabolic network model, the transformation method utilizes the genome scale metabolic network model to predict special structural genes in an FK506 bacterial strain secondary approach gene cluster, the production level of bacterial strains after transformation is improved by 20 percent to 90 percent, the special structural genes in the gene cluster are augmented to improve production capacity, and large application value is achieved in secondary approach rational transformation of microorganism immunosuppressor production bacterial strains. The high-efficiency and systematic method is provided for optimizing of the bacterial strains.
Owner:TIANJIN UNIV
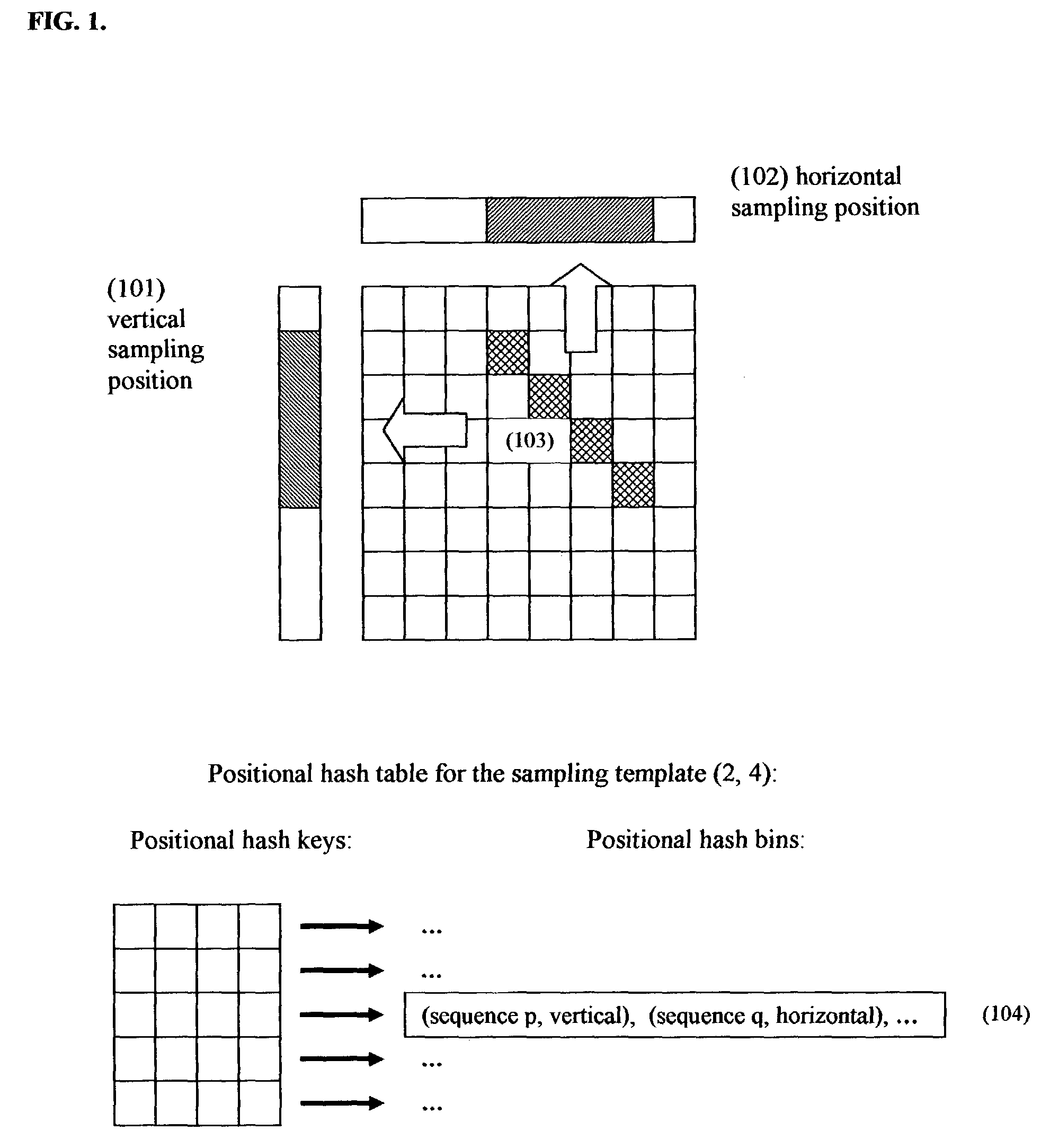
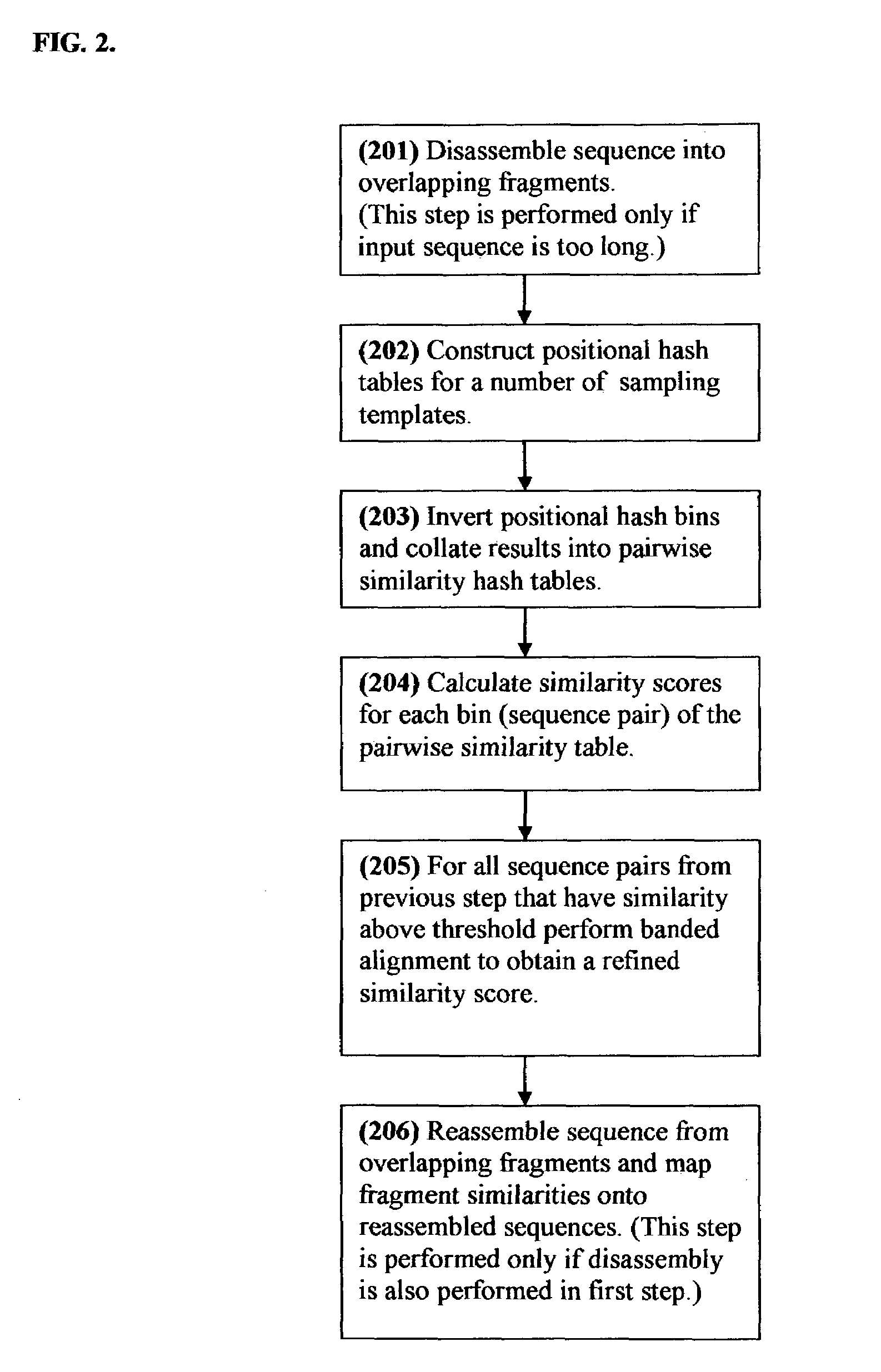
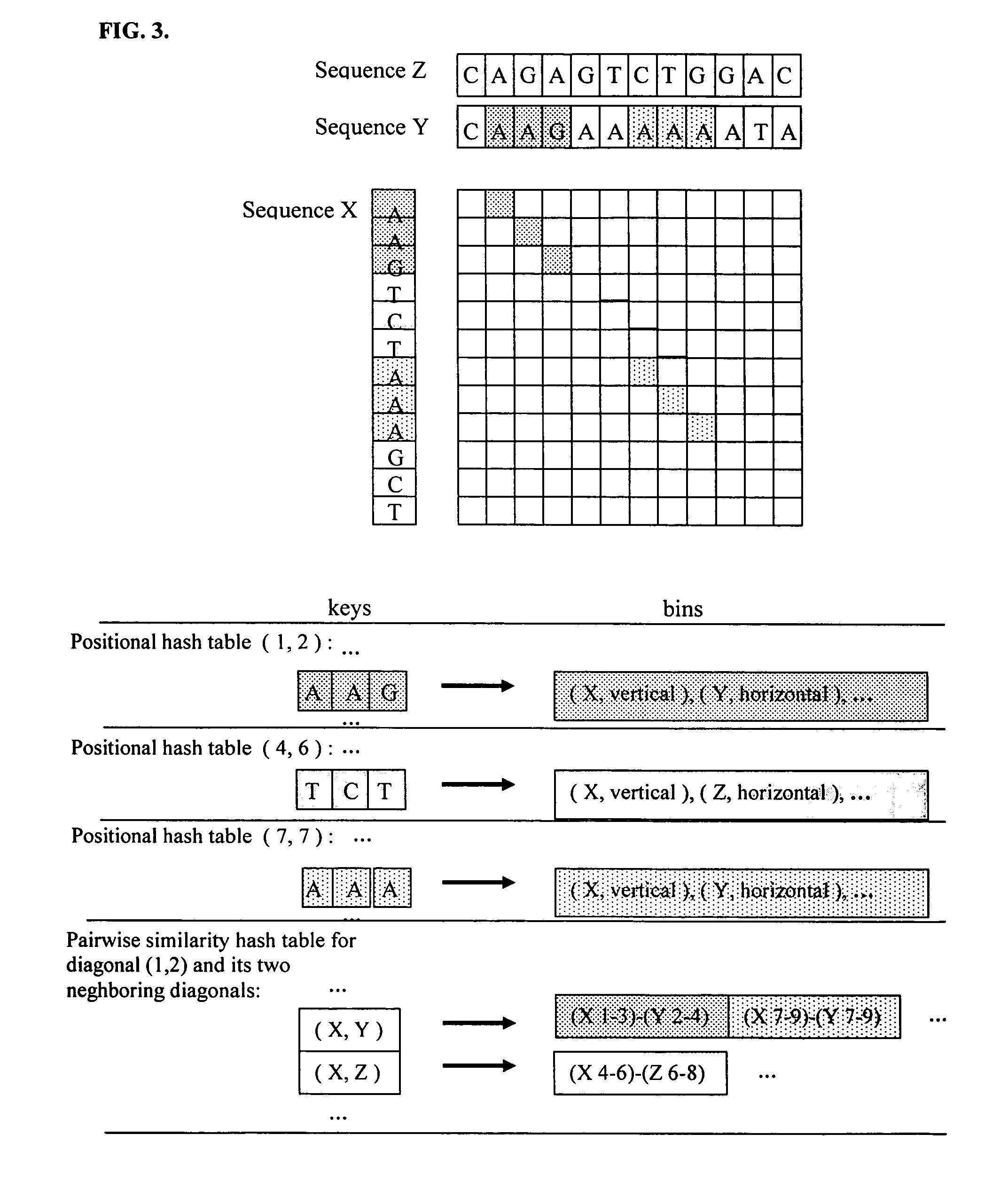
Positional hashing method for performing DNA sequence similarity search
Positional Hashing, a novel method for detecting similarities between texts such as DNA sequences, amino acid sequences, and texts in natural language is disclosed. The method is particularly well suited for large-scale comparisons such as that of mutual comparisons of millions of sequence fragments that result from mammalian-scale sequencing projects and for whole-genome comparisons of multiple mammalian genomes. Positional Hashing is carried out by breaking the sequence comparison problem along its natural structure, solving the subproblems independently, and then collating the solutions into an overall result. The decomposition of the problem into subproblems enables parallelization, whereby a large number of nodes in a computer cluster or a computer farm are concurrently employed on solving the problem without incurring the quadratic time performance penalty characteristic of prior art. Positional Hashing may be used by itself or as a filtering step in conjunction with a variety of sequence comparison algorithms.
Owner:IP GENESIS
Two step genome comparison method for designing quantitative PCR specific primer of guizhouense NJAU 4742
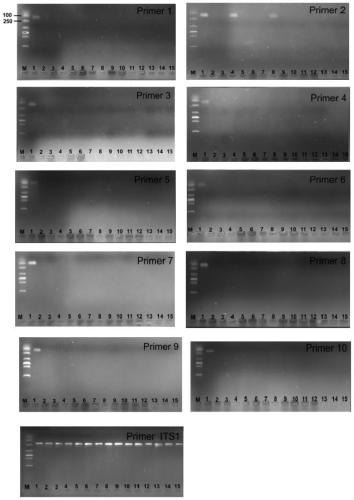
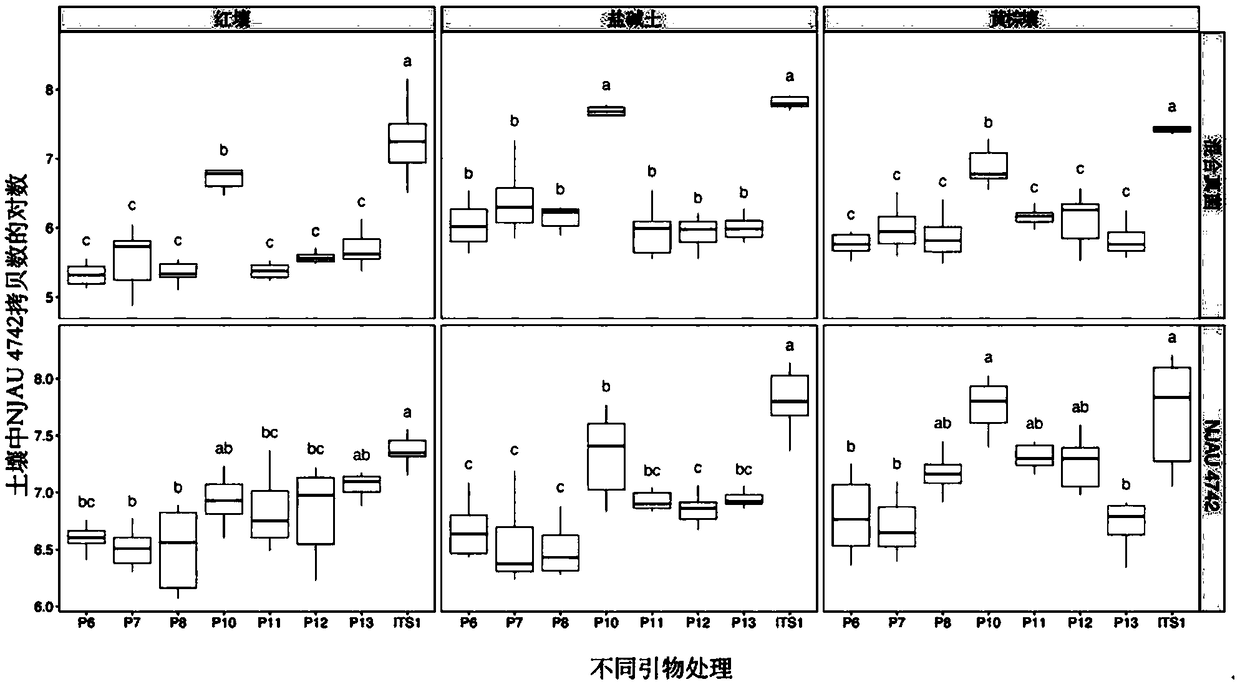
ActiveCN108949908AImprove accuracyMicrobiological testing/measurementMicroorganism based processesGenome alignmentStrain specificity
The invention discloses a two step genome comparison method for designing a quantitative PCR specific primer of guizhouense NJAU 4742. At first, full genome sequence of NJAU 4742 is compared in NCBI database and JGI database; genome segments, which are not matched and greater than 500 bp, are picked out; then the screened segments are compared with the full genome of a guizhouense strain 916; anda primer sequence with unique bases is designed. At the same time, three pairs of primers, which are prepared by inserting labeled segments into a gfp labeled strain (gfp-NJAU 4742), are used to examine the specificity of the primers designed based on the genome. By examining the amplification specificity of strains of same genus and different species, re-detecting the soil addition, and actuallydetecting the potting soil, an optimal quantitative PCR primer is screened out. The provided primer design method and design strain specific primer can be applied to quantitative PCR specific primer design of other strains and quantifying of guizhouense NJAU 4742.
Owner:NANJING AGRICULTURAL UNIVERSITY +1
Automated analysis method for prokaryotic transcriptome based on second generation sequencing
PendingCN110047560AConvenient error queryQuick check for errorsProteomicsGenomicsGenome alignmentStructure analysis
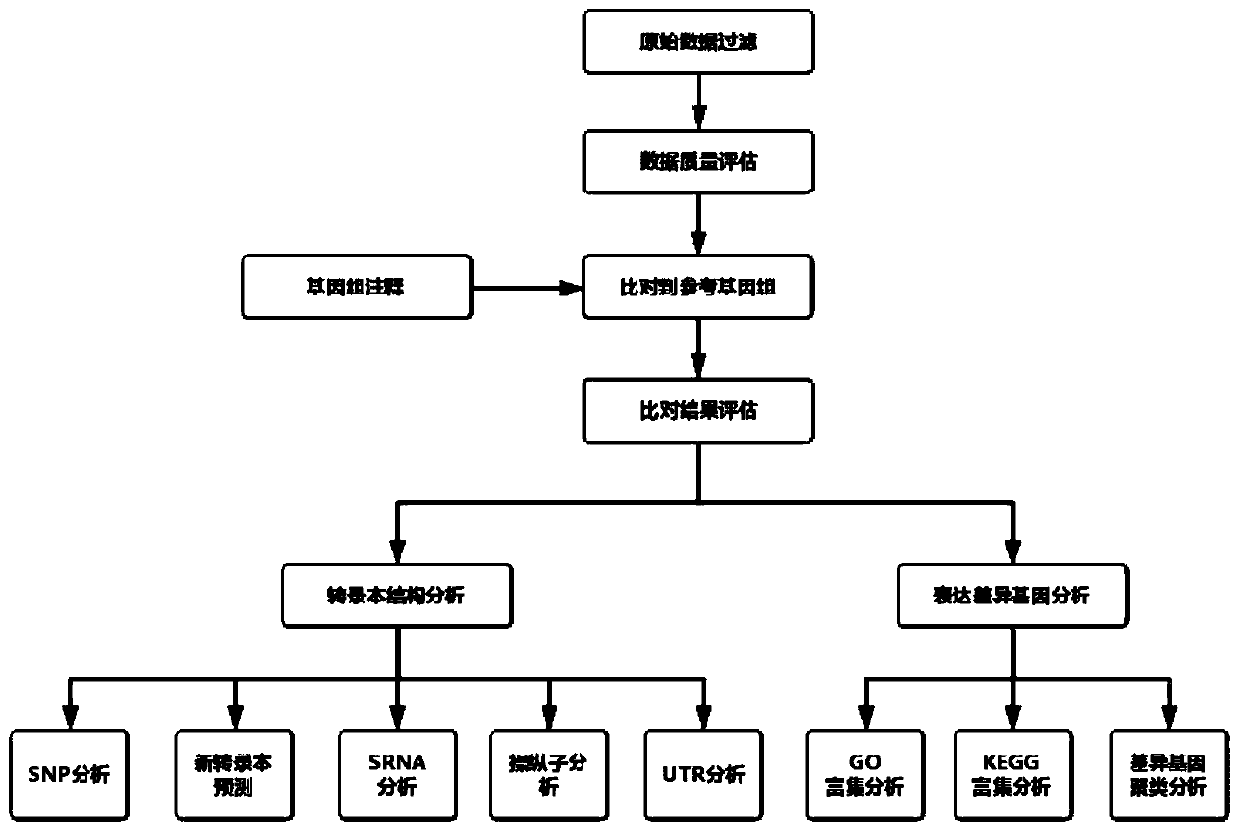
The invention discloses an automated analysis method for prokaryotic transcriptome based on second generation sequencing. The automated analysis method includes the steps: an original logout data filtering and quality control step; a genome comparison step; a transcript structure analysis step; a gene expression quantification and differential expression analysis step; and a result sorting step. The automated analysis method for prokaryotic transcriptome based on second generation sequencing has the advantages of: covering most of the analysis content required by the market, automatically sorting out all the analysis results, and after completing the analysis of each part, automatically counting, visualizing, and sorting the results, so that the results are arranged in order and can be directly used for generation of a report; being visible for all the operation steps, thus being convenient for error query; and when performing each step analysis, recording the used command lines and parameters and the log results generated during the operation, and once the program runs incorrectly, being able to quickly check the error.
Owner:南京派森诺基因科技有限公司
Method for quickly acquiring comparison result data of target genome region
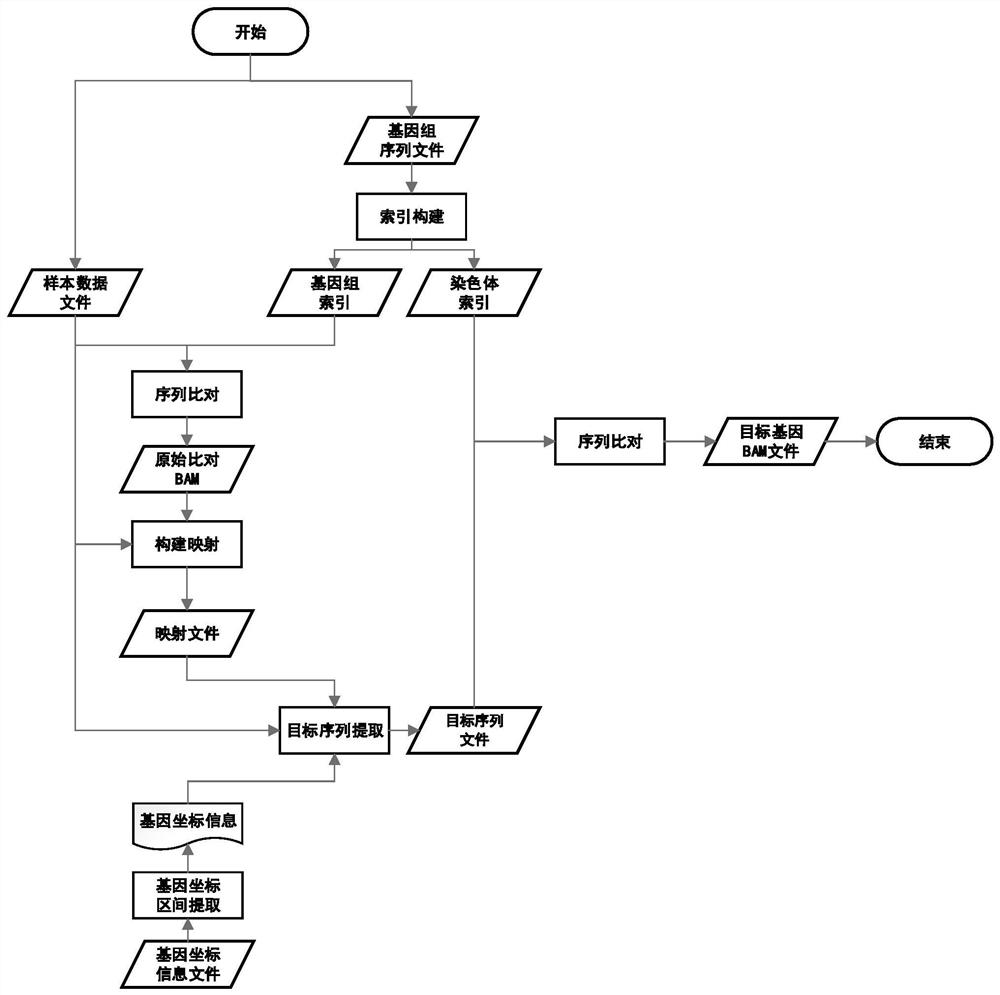
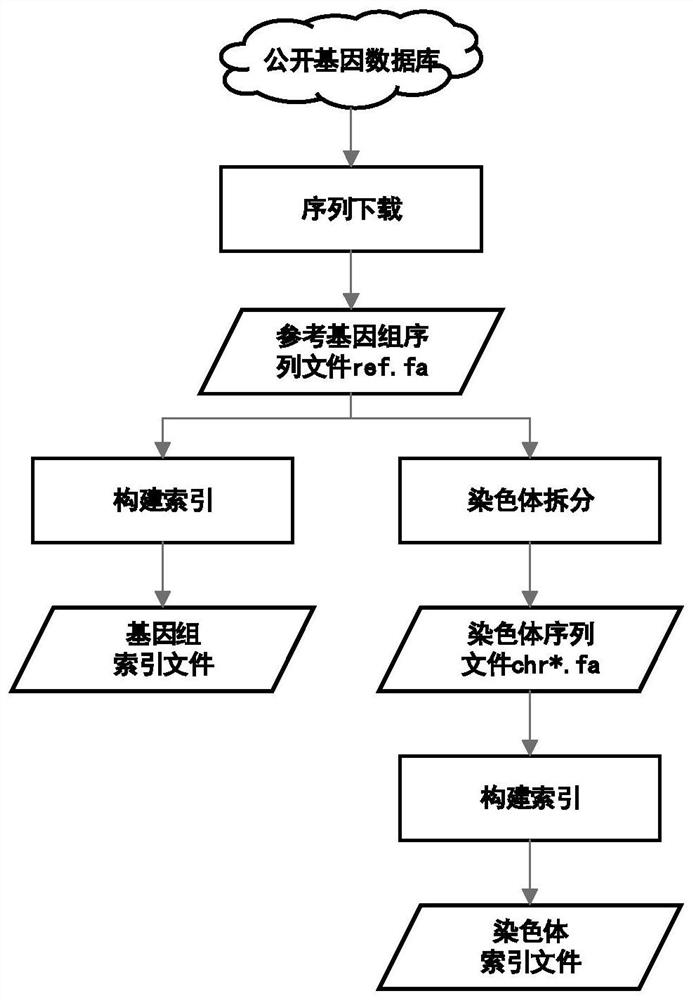
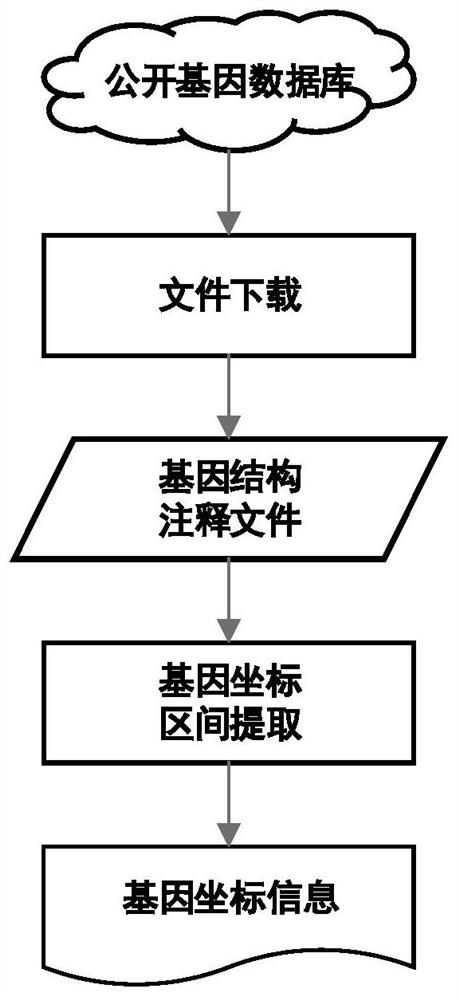
PendingCN113488106AQuick extractionQuick comparisonSequence analysisSpecial data processing applicationsReference genome sequenceTarget gene
The invention discloses a method for quickly acquiring comparison result data of a target genome region. The method comprises the following steps: acquiring a reference genome sequence file and coordinate information files of all genes by using a public genome database on the basis of sample original sequencing data, and constructing a reference genome index file and a chromosome index file; constructing a mapping relationship between the sequence line number of the sample original sequencing data and genome comparison coordinates, and quickly reconstructing sample original sequencing data of a target gene sequence by using the mapping relationship; and carrying out sequence comparison by utilizing the chromosome index file and the sample original sequencing data of the target gene sequence to obtain an original comparison data file of the target gene sequence, and carrying out sequencing and duplicate removal to obtain final comparison result data of a target genome region. The method has the characteristics of simplicity in deployment, convenience in operation, high efficiency, high throughput and wide application range. Compared with an original secondary data BAM file, the obtained result basically has no information loss.
Owner:SUZHOU SMK GENE TECH LTD
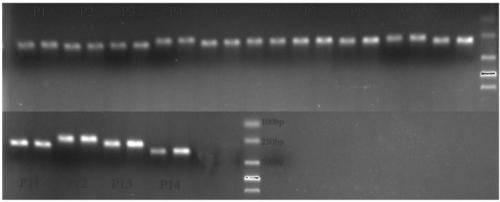
Molecular marker primer composition for identifying inter-hybrid seeds of pears and apples and application thereof
ActiveCN109486991AIncrease credibilityImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationPEARGenome alignment
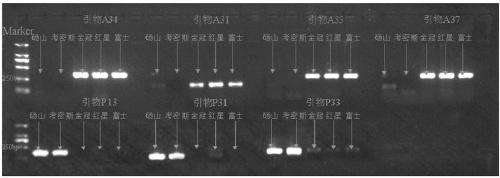
The invention discloses a molecular marker primer composition for identifying inter-hybrid seeds of pears and apples and an application thereof. Specific sequences of pear and apple genomes are detected by a method of whole genome comparison, seven pairs of molecular marker specific primers P13, P31, P33, A31, A33, A34 and A37 are developed newly, and positive and negative primer sequences are asshown in a table 1. By identifying the inter-hybrid offsprings of pears and apples by means of the specific primers, specific amplified strips of the pears, apples and hybrid offsprings can be obtained, separately, that is, the inter-hybrid seeds of the pears and the apples are detected truly. The molecular marker based on the specific sequences of the genomes can be used for identifying the inter-hybrid seeds of pears and apples and has important theoretical and practical guiding meaning in improving genetic breeding progress of fruit trees and accelerating molecular assisted selective breeding.
Owner:NANJING AGRICULTURAL UNIVERSITY
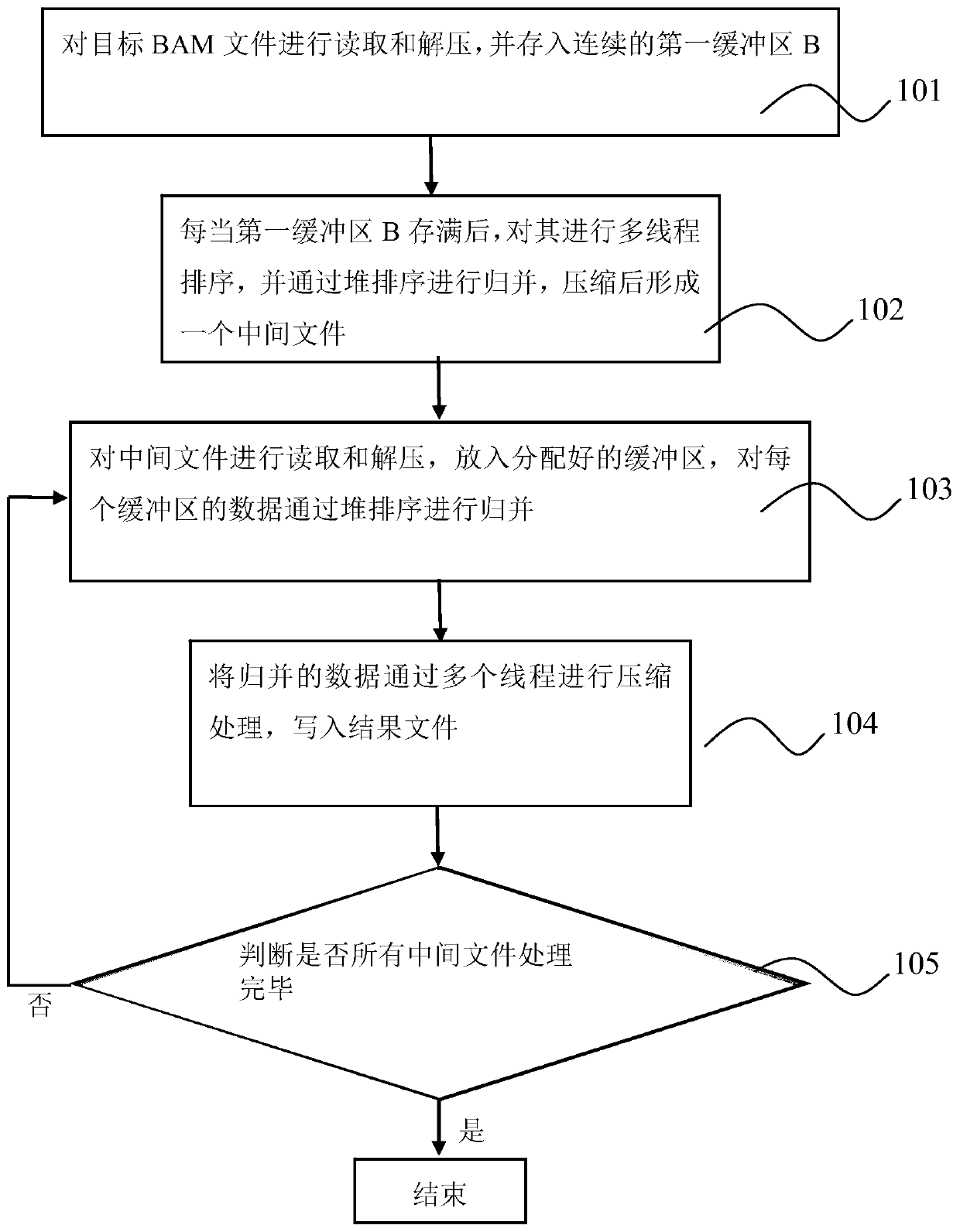
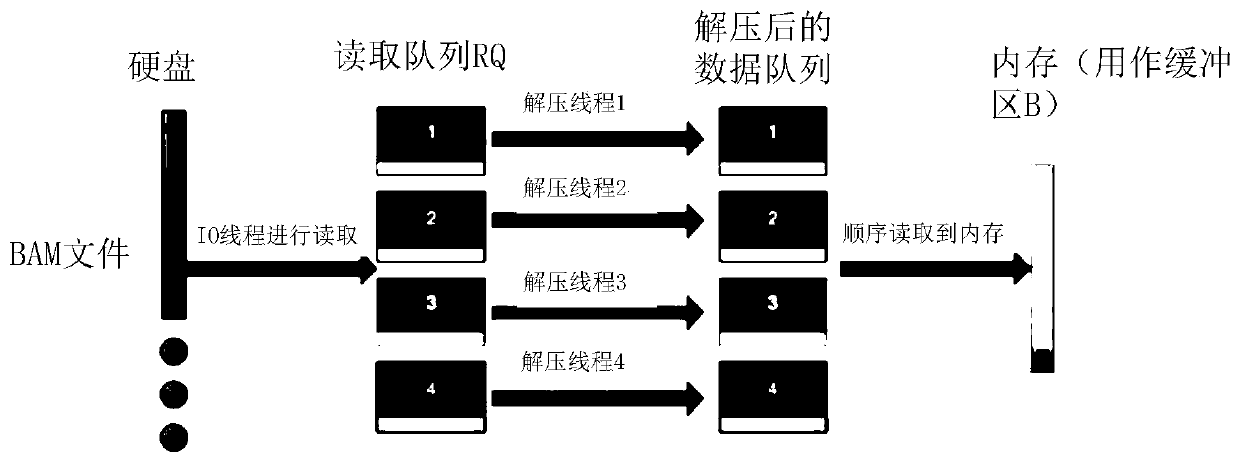
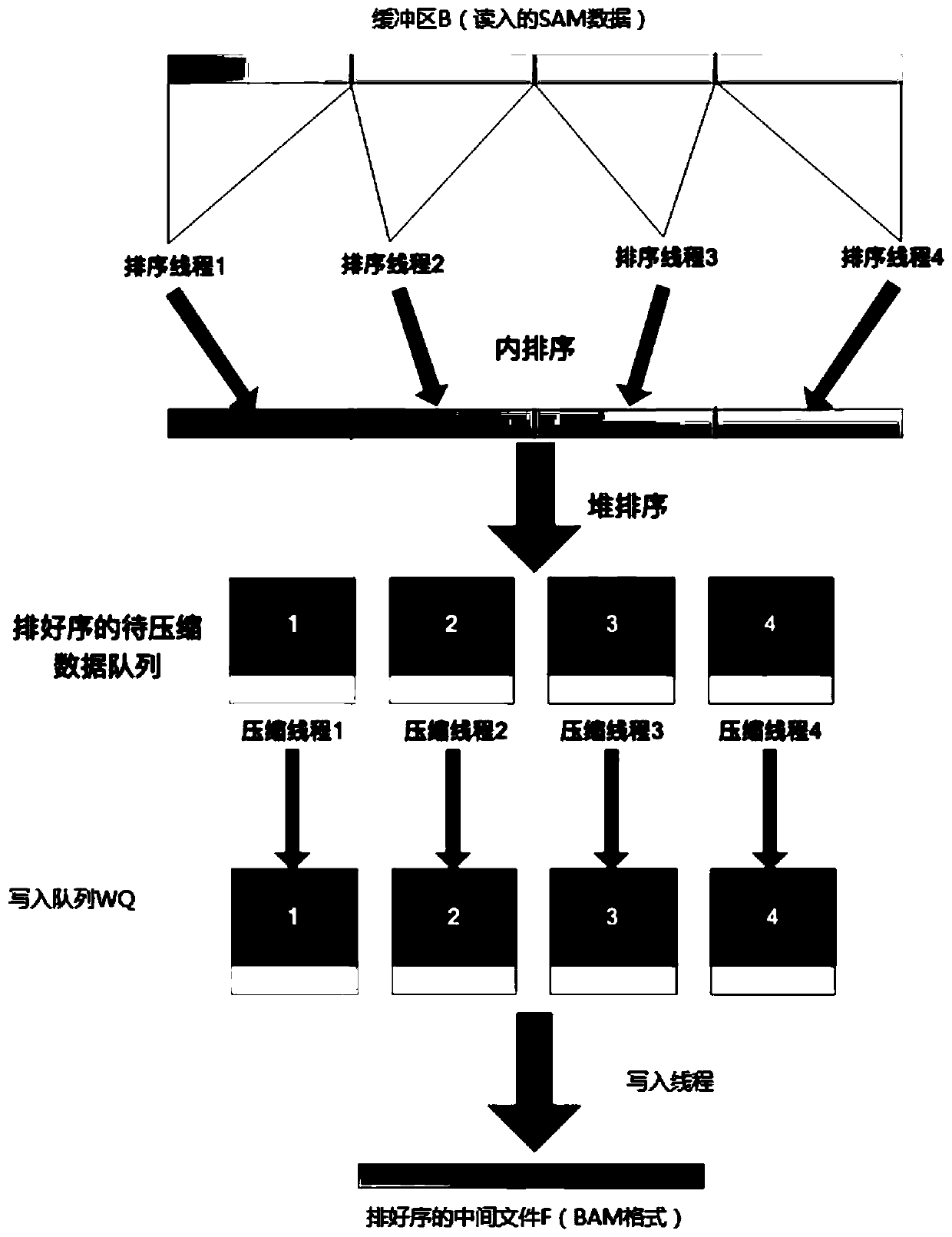
Parallel acceleration method for sequencing big data genome comparison files
PendingCN110767265AImprove reading and writing efficiencyReduce in quantitySequence analysisInstrumentsParallel computingHeapsort
The invention discloses a parallel acceleration method for sequencing big data genome comparison files. The method comprises steps of reading and decompressing a target BAM file, and storing the target BAM file into a continuous first buffer area B; performing multi-thread sorting after the first buffer area B is full, performing merging through heap sorting to form intermediate files; reading theintermediate files in sequence, putting the intermediate files into an associated second buffer area MB, and merging the data of each second buffer area MB through heap sorting; compressing the merged data through a plurality of threads, and writing the compressed data into a result file. The method is advantaged in that the threads are independently distributed for reading and decompressing, thread pools are constructed for decompressing and compressing respectively, the number of developed threads is reduced, multi-thread resources are fully utilized, file reading and writing efficiency isimproved, the number of intermediate files is reduced, the number of times of memory copying operation is reduced, and the processing time is shortened.
Owner:INST OF COMPUTING TECH CHINESE ACAD OF SCI
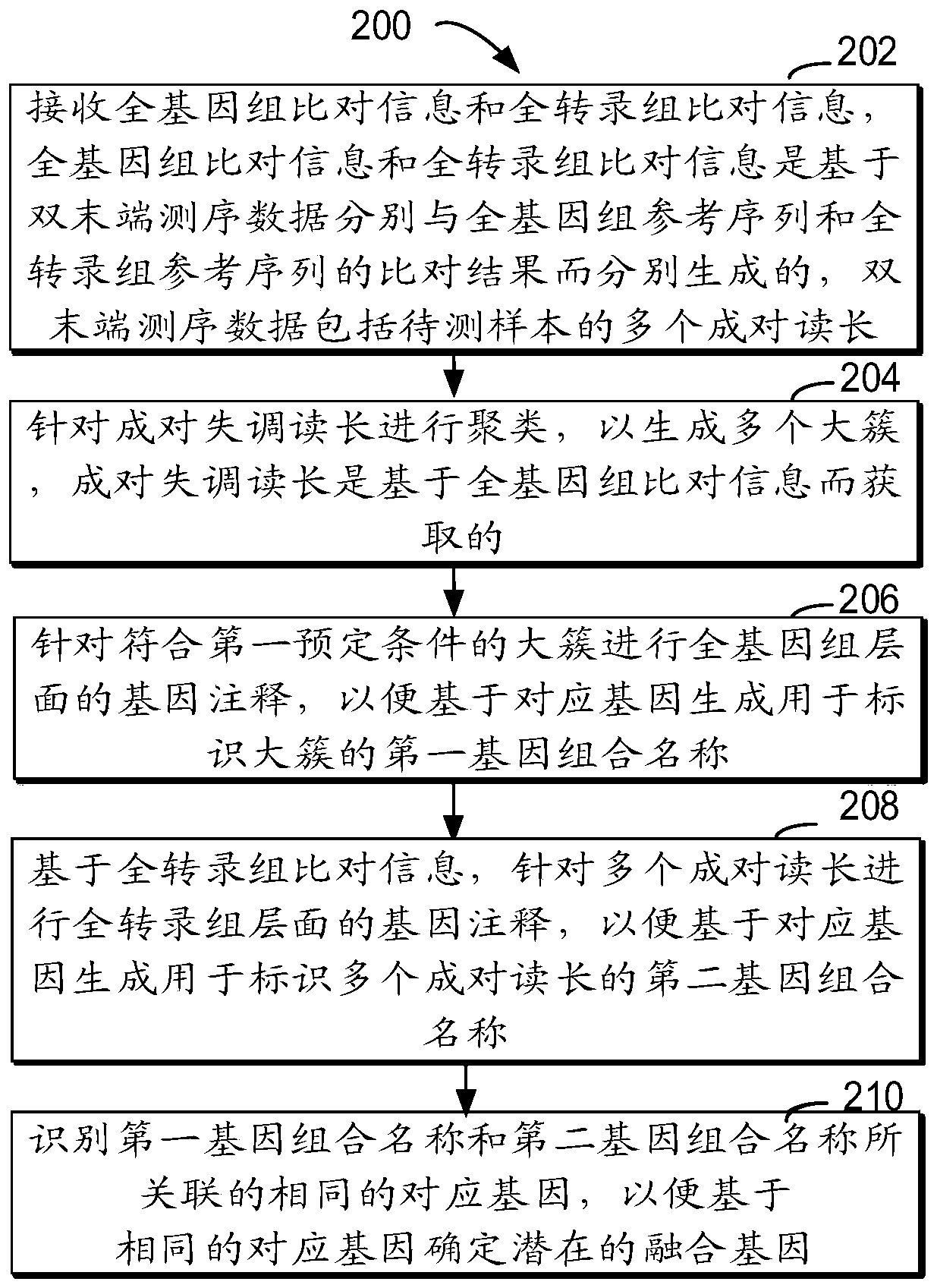
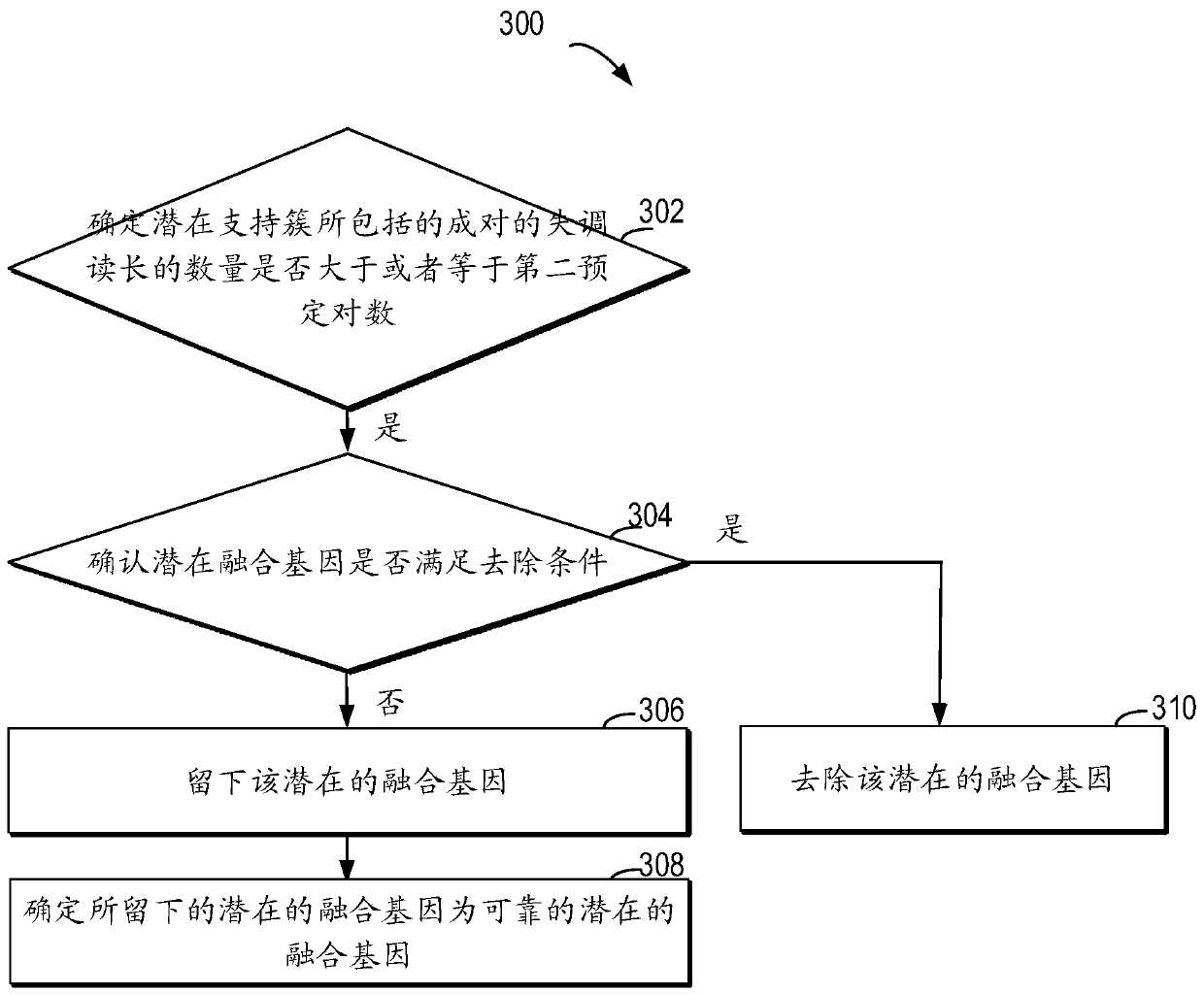
Method for detecting RNA level gene fusion, electronic device, and computer storage medium
ActiveCN111292809AQuick and correct identificationBiostatisticsProteomicsGenome alignmentGene Annotation
The invention relates to a method for detecting RNA level gene fusion, electronic equipment and a computer storage medium. The method comprises the following steps: receiving whole genome comparison information and whole transcriptome comparison information; clustering the paired maladjusted read lengths to generate a plurality of large clusters; carrying out whole-genome-level gene annotation onthe large clusters meeting a first preset condition so as to generate first gene combination names for identifying the large clusters based on the corresponding genes; based on the whole transcriptomecomparison information, performing whole transcriptome-level gene annotation on the plurality of paired read lengths so as to generate a second gene combination name for identifying the plurality ofpaired read lengths based on the corresponding gene; and identifying the same corresponding genes associated with the first gene combination name and the second gene combination name so as to determine the same corresponding genes as potential fusion genes. The method is beneficial to rapid and correct recognition of false positive results, and the false positive results of a gene fusion detectionresult can be obviously reduced.
Owner:SHANGHAI ORIGIMED CO LTD +1
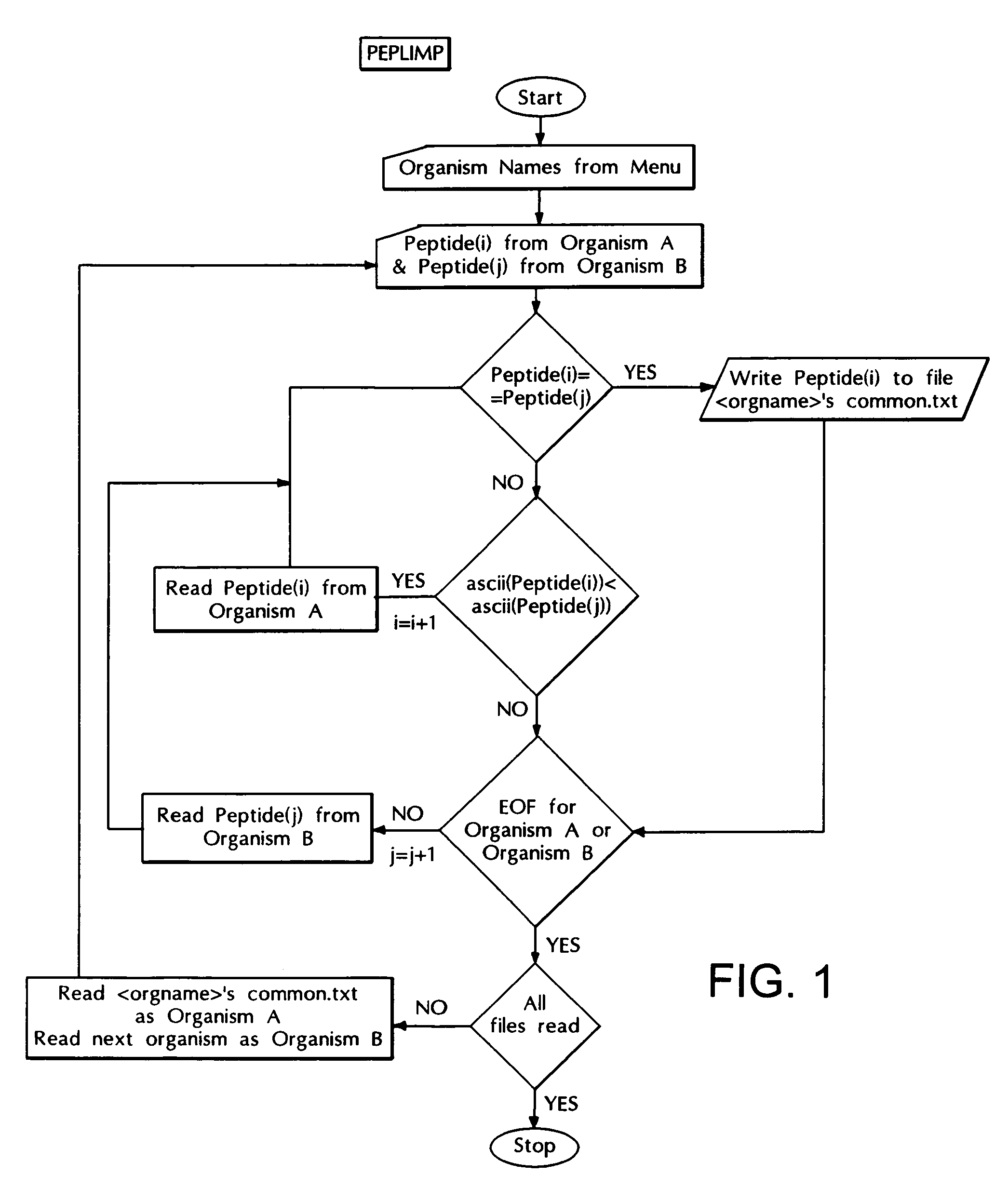
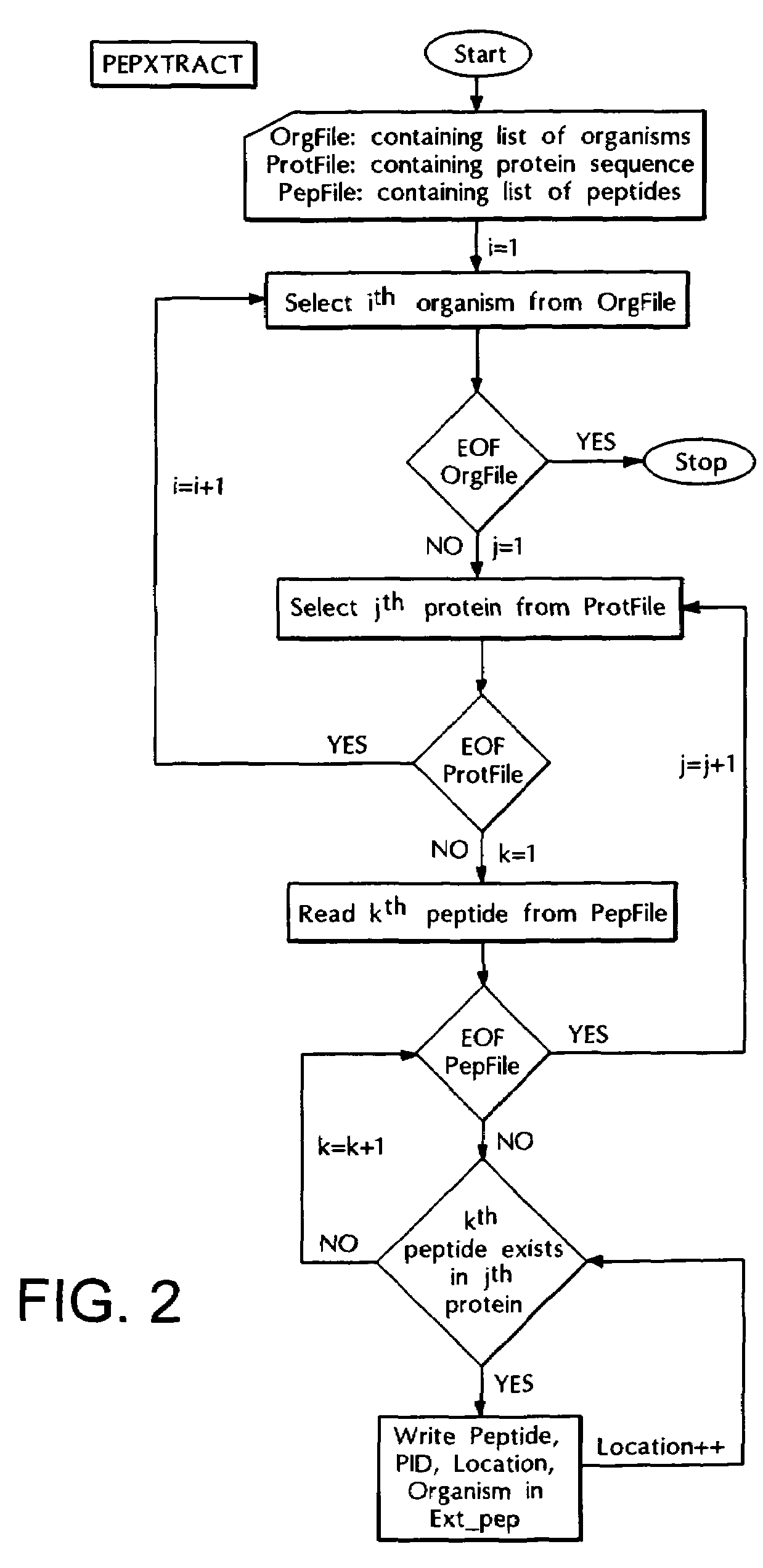
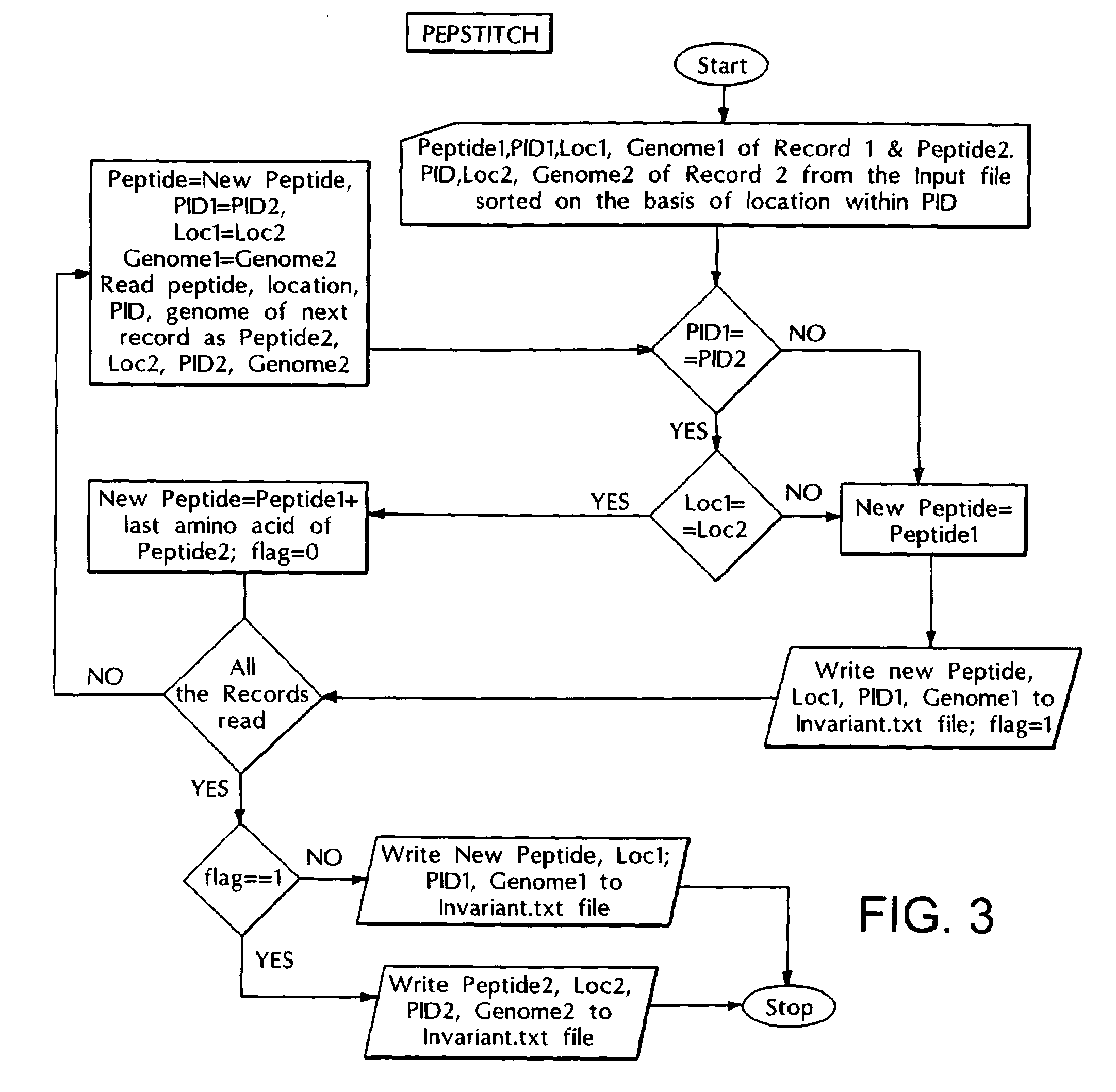
Computer based method for identifying peptides useful as drug targets
InactiveUS7657378B1Accurate distinctionMicrobiological testing/measurementPeptide preparation methodsPresent methodDrug target
The present invention relates to a novel computer based method for performing genome-wise comparison of several organisms, the said computational method involves creation of peptide libraries from protein sequences of several organisms and subsequent comparison leading to identification of conserved invariant peptide motifs, and to this end several invariant peptide motifs have been identified by direct sequence comparison between various bacterial organisms and host genomes without any a priori assumptions, and the present method is useful for identification of potential drug targets and can serve as drug screen for broad-spectrum antibacterials as well as for specific diagnosis of infections, and in addition, for assignment of function to proteins of yet unknown functions with the help of such invariant peptide motif signatures.
Owner:COUNCIL OF SCI & IND RES
Method for rapid screening of garden establishment parent materials of cedar seed garden
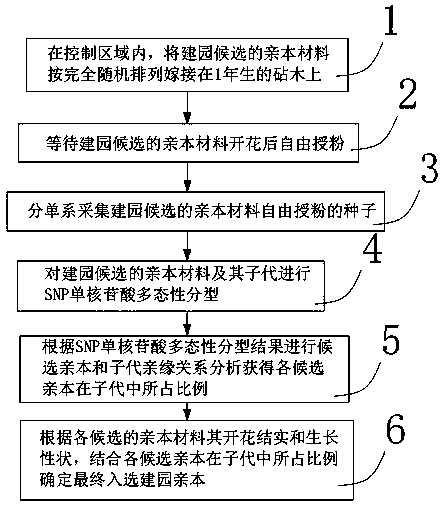
PendingCN111296285AReduce workloadHigh polymorphismGraftingPlant genotype modificationOpen pollinationEcology
The invention provides a method for rapid screening of garden establishment parent materials of a cedar seed garden. The method comprises the following steps: conducting open pollination on candidateparents of a garden establishment region in a control area; obtaining a filial generation of each parent after the open pollination through monophyletic seed collection; using an SNP molecular markerfor genotyping of the parents and the filial generations so as to determine the parents of which the filial generations are dominant; and determining the parent materials for garden establishment according to traits of blossom and growth. Forest trees have large genomes in general. The method rapidly detects characteristics of differences of genomes among forest plant individuals with the SNP molecular marker and is characterized by fast screening, a large processing amount, high polymorphism, high stability and a wide coverage scope.
Owner:GUANGDONG ACAD OF FORESTRY
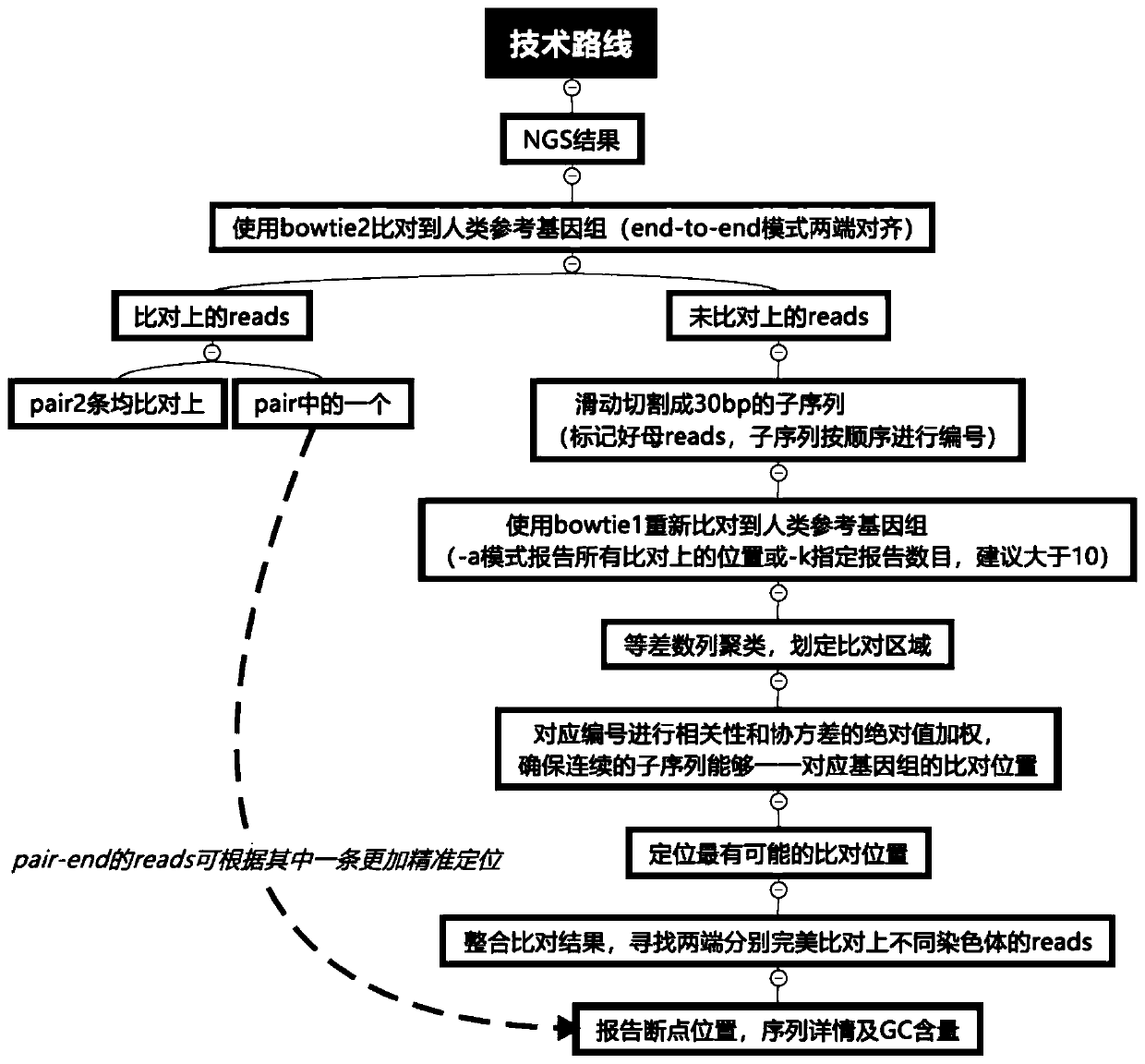
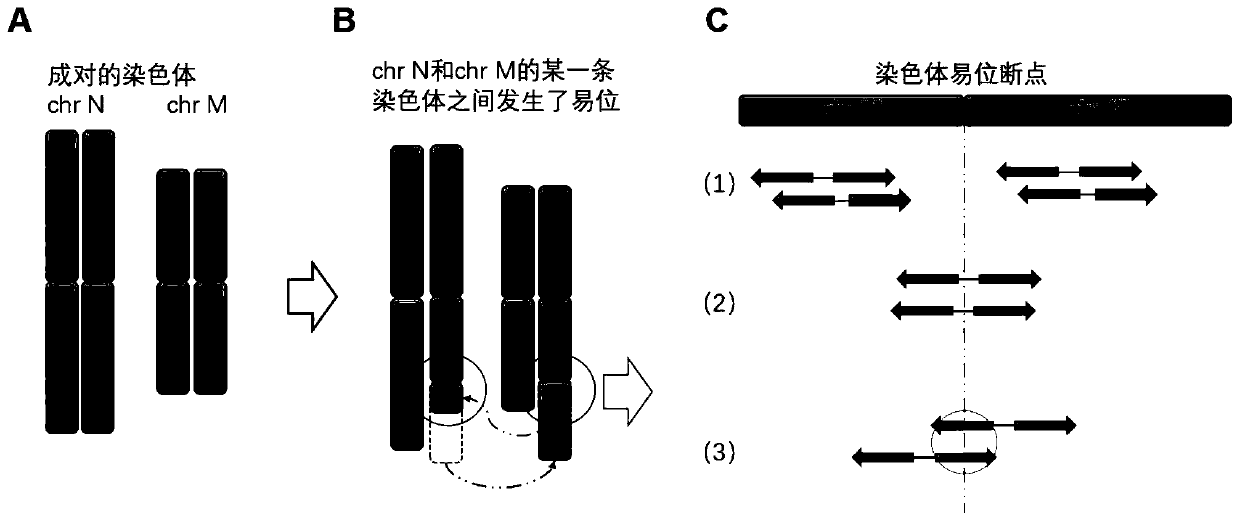
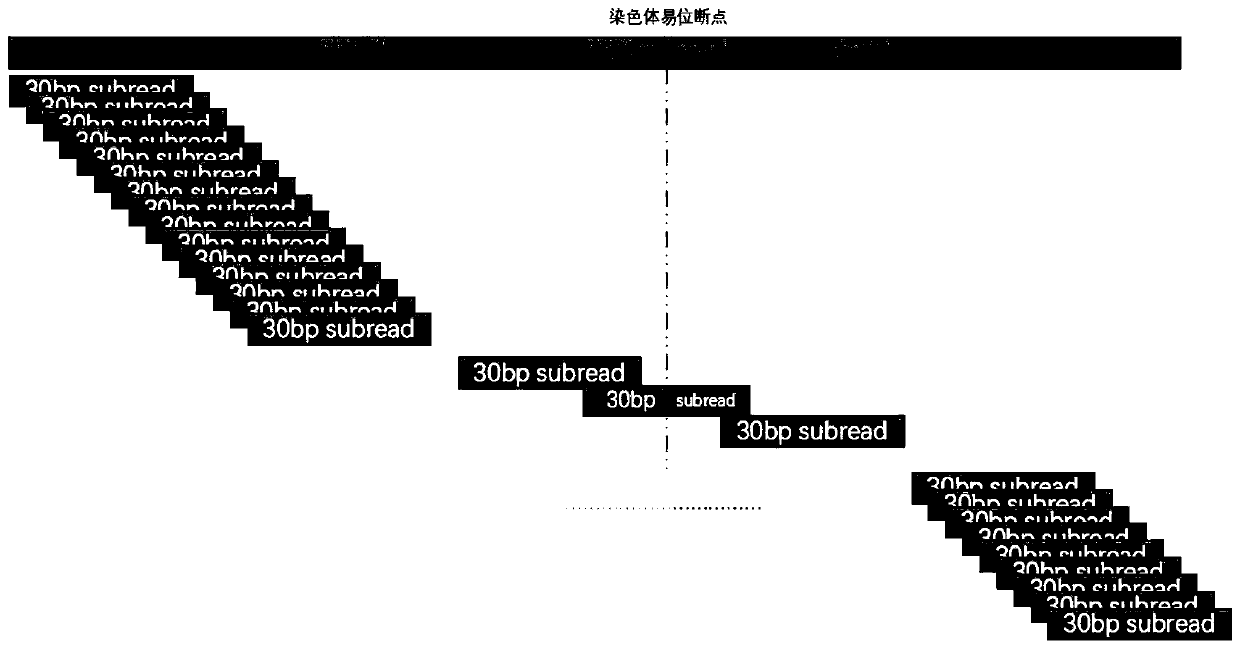
NGS-based chromosome balanced translocation detection and analysis system and its application
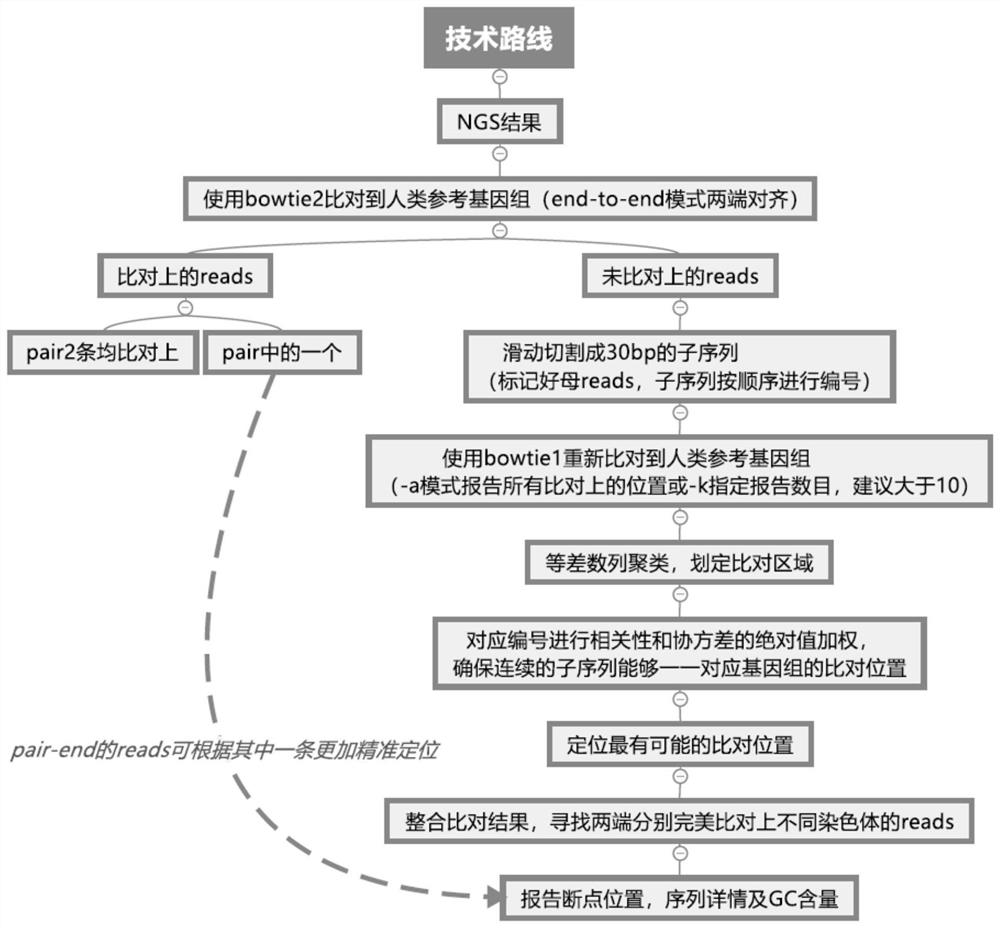
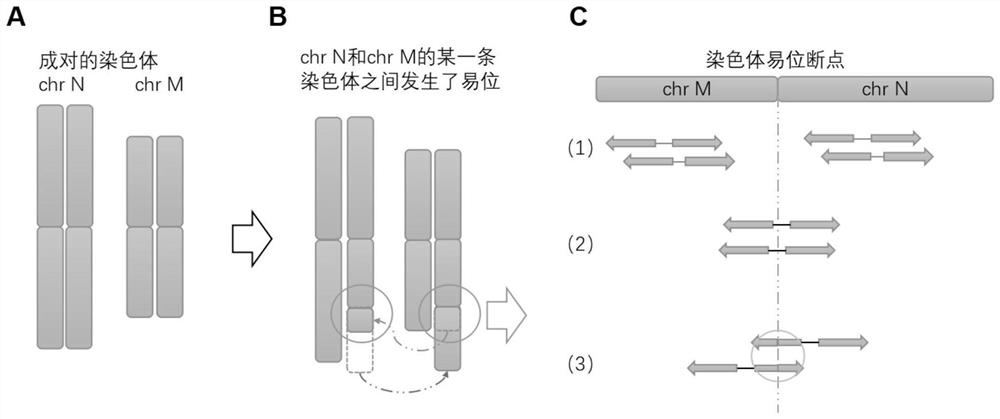
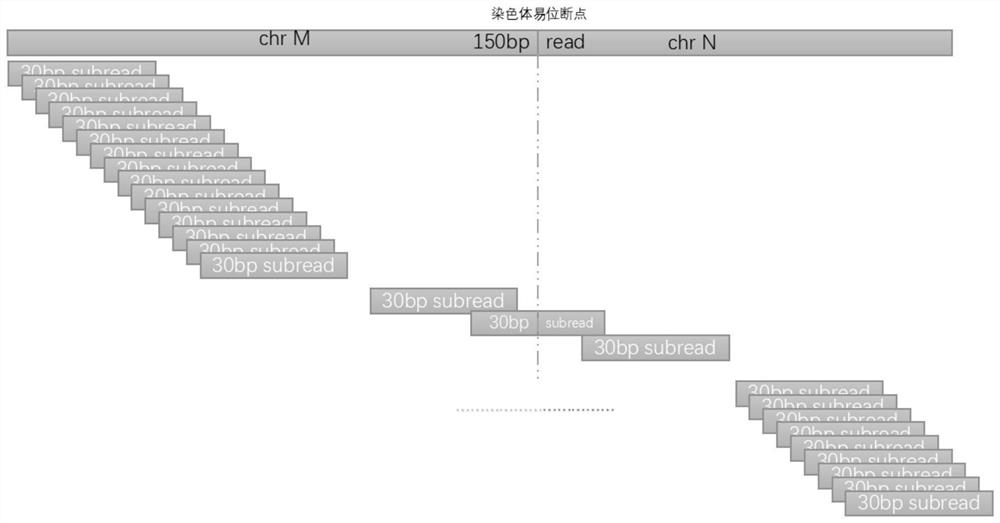
ActiveCN111276189BQuick analysisImprove analytical precisionSequence analysisInstrumentsHuman DNA sequencingGenome human
The invention relates to an NGS-based chromosome equilibrium translocation detection and analysis system and application, and belongs to the technical field of gene detection biological information. The detection and analysis system comprises a data acquisition module, a genome comparison module, a sliding cutting module, a short sequence comparison module, a breakpoint analysis module and a result output module. Firstly, reads which cannot be simultaneously compared to a human reference genome at two ends are screened out and cut into continuous short sub-sequences by sliding; the sub-sequences are compared to the human reference genome again, so that the reads sequence originally comprising balanced translocation points can be compared with the human genome through short fragments aftercutting; and through clustering grouping of comparison positions and a correlation covariance weighting algorithm of corresponding sub-sequence numbers, reads of which the two ends are respectively compared with different chromosomes can be quickly screened out, and a balanced translocation breakpoint is accurately positioned, so that a sequence designed by a primer is provided for subsequent Sanger sequencing verification. The application range comprises NIPT, PGS, PGD, common NGS sequencing results and the like.
Owner:GUANGZHOU KINGMED TRANSFORMATIVE MEDICINE INST CO LTD
NGS-based chromosome equilibrium translocation detection and analysis system and application
ActiveCN111276189AQuick analysisSimple stepsSequence analysisInstrumentsHuman DNA sequencingGenome human
The invention relates to an NGS-based chromosome equilibrium translocation detection and analysis system and application, and belongs to the technical field of gene detection biological information. The detection and analysis system comprises a data acquisition module, a genome comparison module, a sliding cutting module, a short sequence comparison module, a breakpoint analysis module and a result output module. Firstly, reads which cannot be simultaneously compared to a human reference genome at two ends are screened out and cut into continuous short sub-sequences by sliding; the sub-sequences are compared to the human reference genome again, so that the reads sequence originally comprising balanced translocation points can be compared with the human genome through short fragments aftercutting; and through clustering grouping of comparison positions and a correlation covariance weighting algorithm of corresponding sub-sequence numbers, reads of which the two ends are respectively compared with different chromosomes can be quickly screened out, and a balanced translocation breakpoint is accurately positioned, so that a sequence designed by a primer is provided for subsequent Sanger sequencing verification. The application range comprises NIPT, PGS, PGD, common NGS sequencing results and the like.
Owner:GUANGZHOU KINGMED TRANSFORMATIVE MEDICINE INST CO LTD
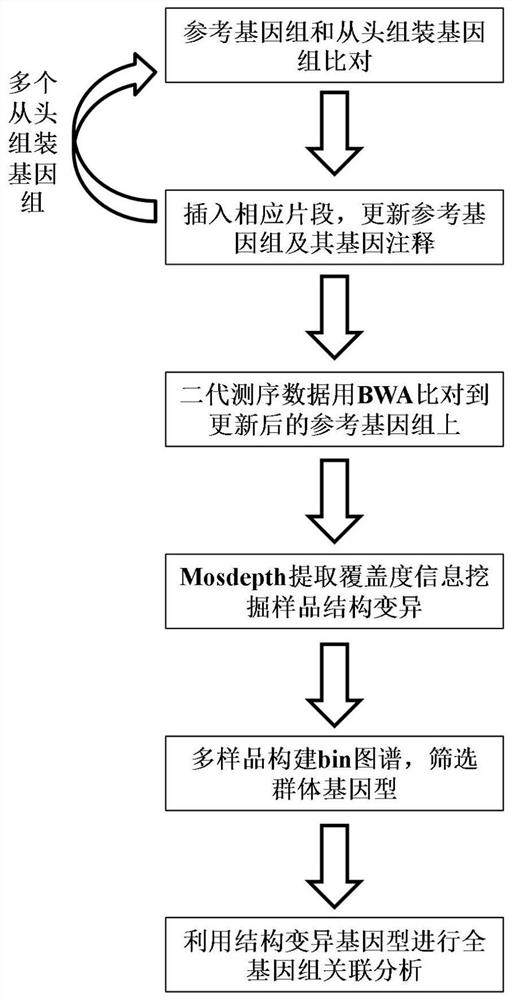
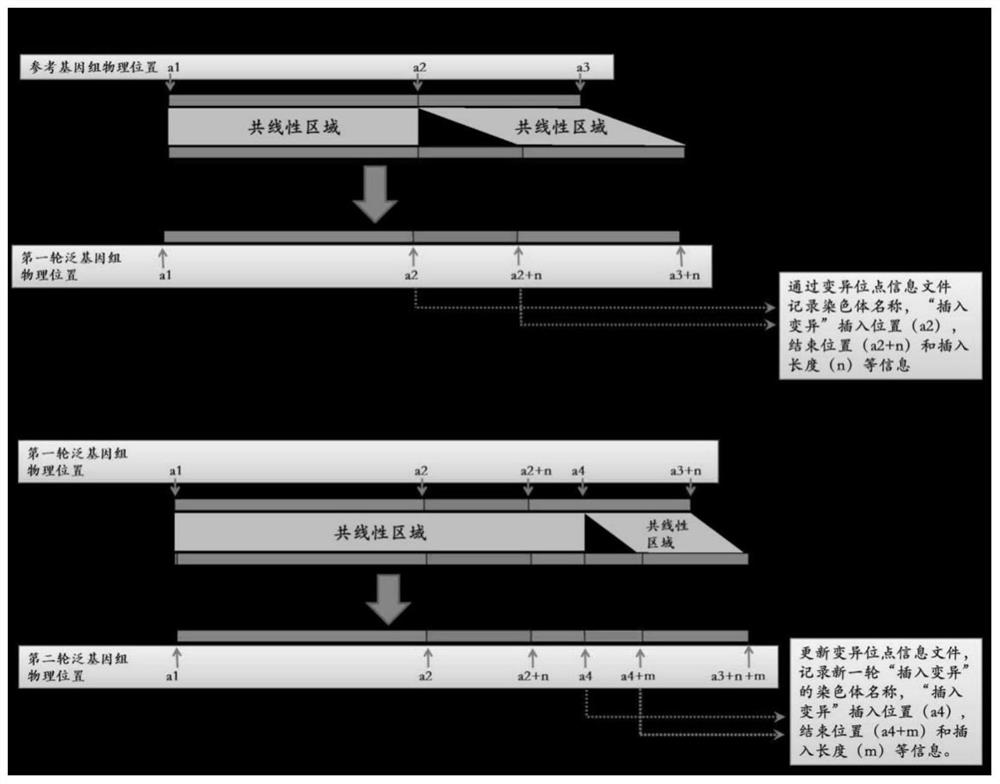
Whole genome association analysis method based on comparison of multiple genomes and next-generation sequencing data
ActiveCN113628685AImplementation of association analysisEasy to understandProteomicsGenomicsGene PositionWhole Genome Association Analysis
The invention discloses a whole genome association analysis method based on comparison of multiple genomes and next-generation sequencing data, which comprises the following steps of: 1, comparing a reference genome with a de novo assembly genome file by using comparison software; and updating the annotation gene positions or structures in the reference genomes; 3, if a plurality of de novo assembly genomes exist, sequentially iteratively updating the reference genomes; 4, comparing the next-generation sequencing data of the samples to the updated reference genome by using comparison software; 5, collecting demarcation point position information of all structural variations of all the samples into a set, constructing a population genotype; and 6, performing functional gene candidate according to the association site and the updated reference genome annotation file. According to the method, the whole genome association analysis is performed by using the structure variation genotype data.
Owner:RICE RES INST GUANGDONG ACADEMY OF AGRI SCI
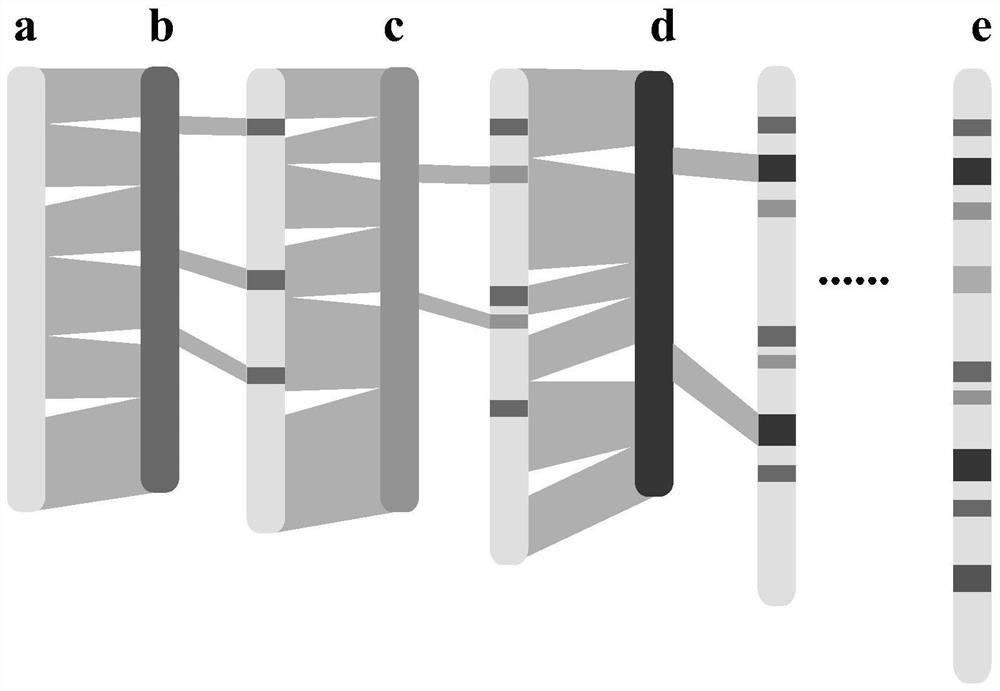
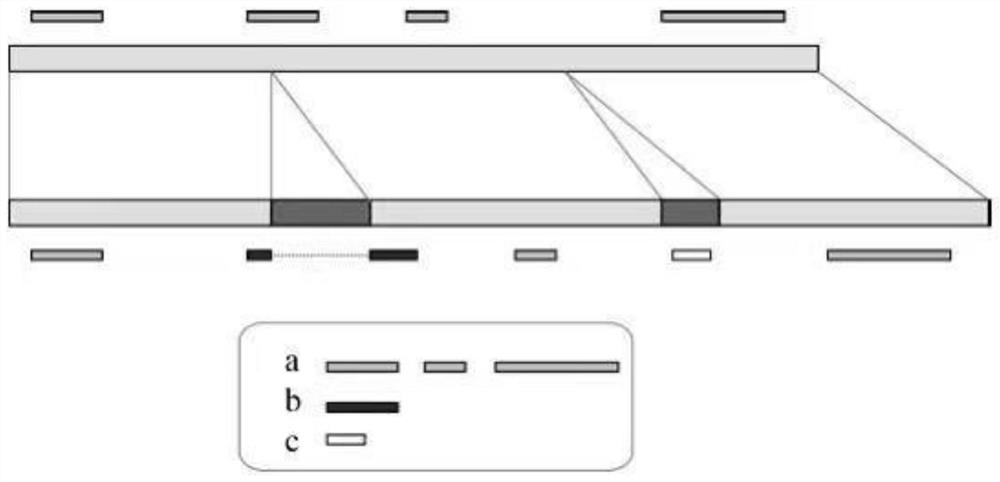
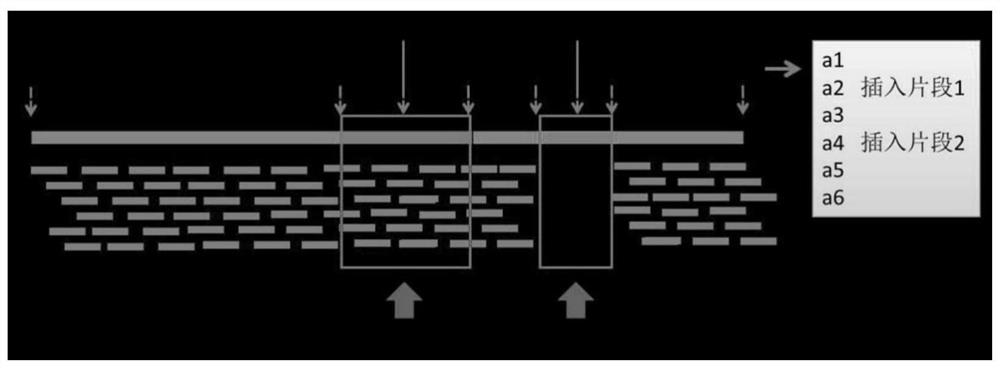
Generic genome construction method and corresponding structural variation mining method thereof
ActiveCN113571131AQuality improvementAchieve large-scale applicationBiostatisticsProteomicsTheoretical computer scienceEngineering
The invention belongs to the technical field of genome data analysis, and particularly relates to a generic genome construction method and a corresponding structural variation mining method thereof, structural variation obtained through genome comparison is put back to a linear genome, meanwhile, a structural variation site information file is added, and an efficient analysis generic genome which is linearized in the form and considers various structure variation forms is constructed. The generic genome not only can capture more brand-new structural variations which are not found by a reference genome, but also better displays the captured structural variations by combining a linearization method with a variation site information file, so that the constructed generic genome is easier to understand and analyze, and is more beneficial to subsequent application; according to the method and the process for analyzing the genome structure variation based on the generic genome and the second-generation sequencing data, a complete program code is compiled, and the efficient and accurate mining of the structure variation based on the relatively low-cost second-generation sequencing data is realized.
Owner:RICE RES INST GUANGDONG ACADEMY OF AGRI SCI
Streptococcus agalactiae attenuated strain YM001 genome sequence feature and use thereof
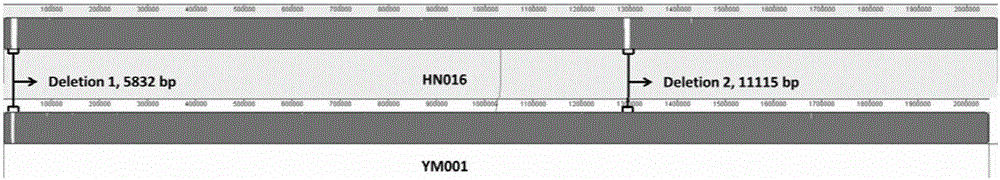
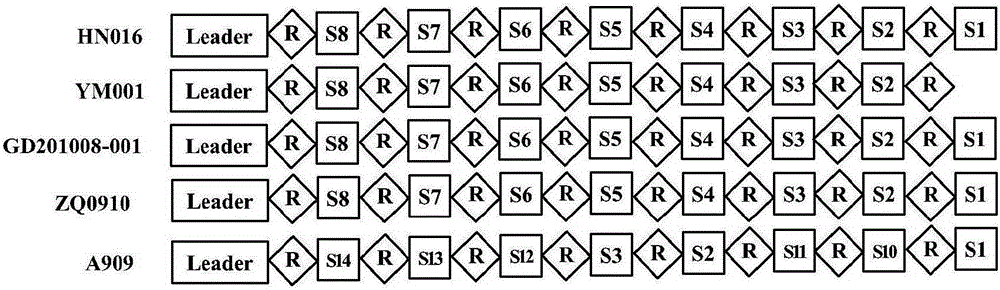
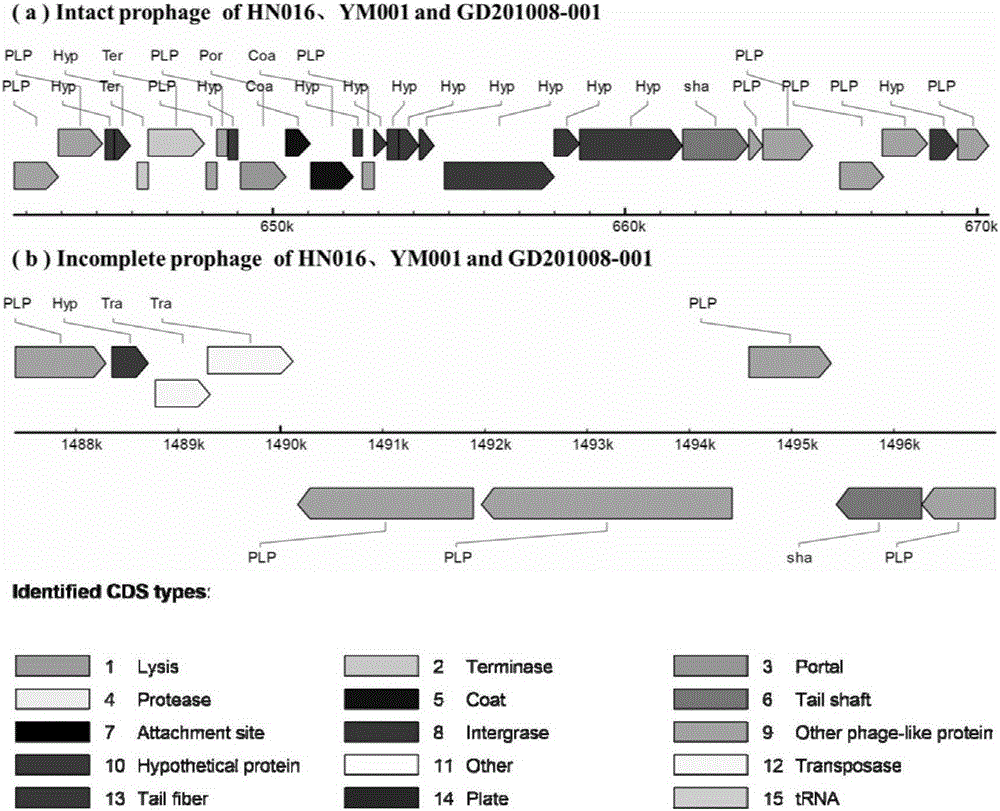
The invention relates to a streptococcus agalactiae attenuated strain YM001 genome sequence feature and a use thereof, especially relates to a streptococcus agalactiae attenuated strain genome sequence feature and concretely relates to streptococcus agalactiae attenuated strain whole genome sequencing, sequence analysis and genome comparison analysis. Compared with a maternal wild virulent strain HN016 genome, the YM001 genome has two large fragment losses, the loss sizes are 5832 bp and 11115 bp, and three rRNA and eleven tRNA gene losses and ten functional gene losses or function damage are caused. Compared with the HN016 genome, the YM001 genome has ten small fragment losses and 28 mononucleotide variants. The YM001 genome variation feature can be used for protection of attenuated strain YM001 intellectual property and construction and screening of a streptococcus agalactiae attenuated strain.
Owner:GUANGXI ACADEMY OF FISHERY SCI
Broccoli SSR (simple sequence repeat) primer based on transcriptome sequencing and development
ActiveCN107604094AHigh polymorphismImprove usabilityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceData information
The invention provides a broccoli SSR (simple sequence repeat) primer based on transcriptome sequencing and development. The broccoli SSR primer comprises 17 pairs of primers, wherein the sequences ofthe primers are shown in SEQ ID NO.1 to 34. The broccoli SSR primer has the advantages that a large amount of transcript data information is obtained by the transcriptome sequencing of the broccoli,a large amount of SSR primers is developed, the developed SSR marker is effectively demonstrated by the representative germplasm resources collected by a topic team, and the foundation is laid for thesubsequent study of genetic diversity, comparison and study of genomes, constructing of genetic spectrums, QTL (quantitative trait loci) location of important characters, utilization and storage of core resources, auxiliary breeding of molecules, and the like of the broccoli.
Owner:CROP RES INST OF FUJIAN ACAD OF AGRI SCI
Construction method of whole-genome exon sequence capture probe
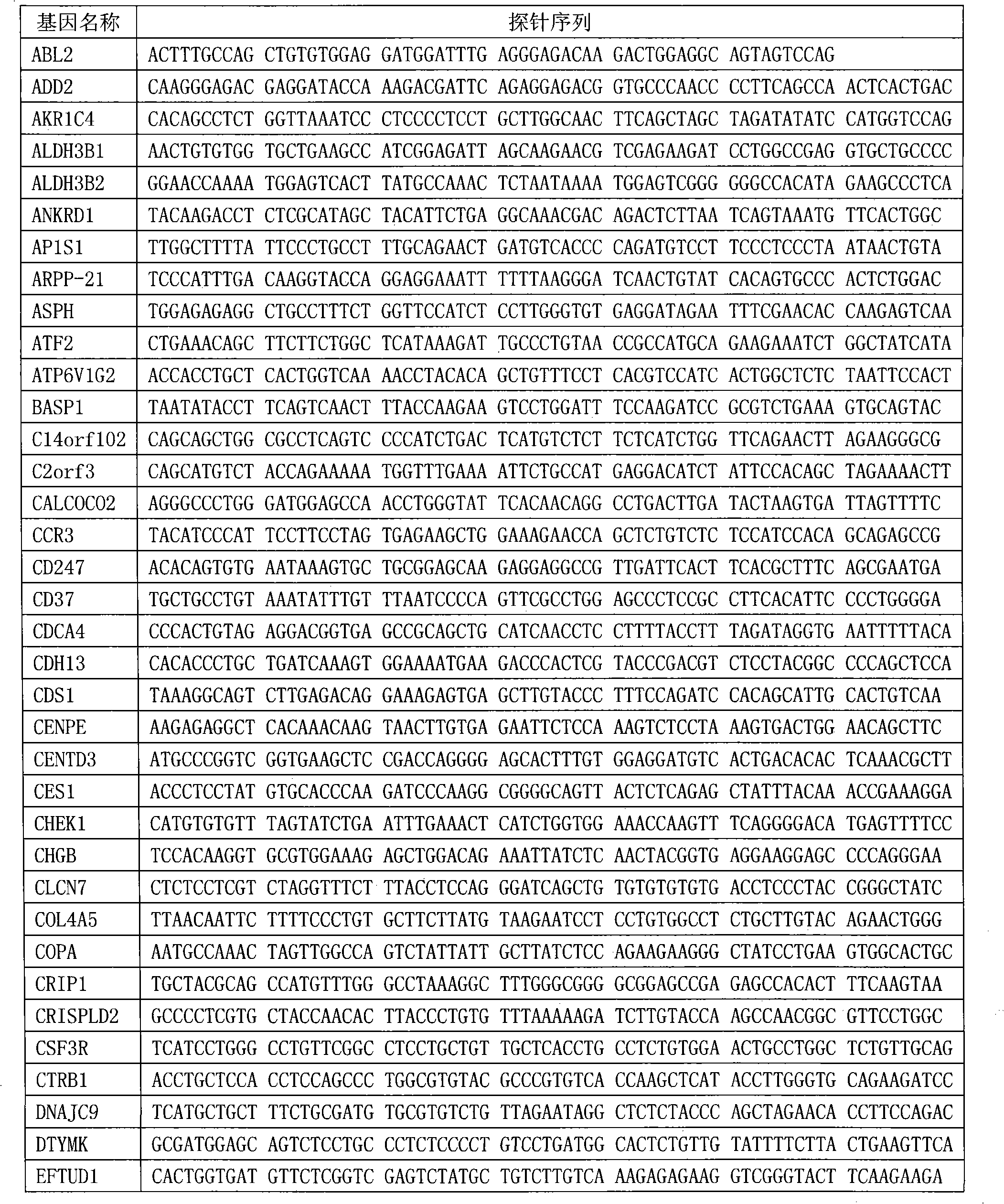
The invention relates to a kit for capturing a whole-genome exon sequence. The kit is a target sequence capture system based on the hybridization principle. An exon capture probe is designed, and a large quantity of exomes are obtained by hybridization to perform high-throughput sequencing and human whole genome comparison; and then, data analysis is performed, and the capture efficiency is calculated. The kit provided by the invention can provide high-quality target sequence enrichment, and has high specificity, high accuracy and excellent reproducibility in whole genome exon sequencing; the whole genome exon sequencing becomes the most effective policy for identifying the disease-causing gene of the Mendel disease; and the kit is also applied to the study and clinical diagnosis of the gene susceptible to complex diseases, and has a wide application prospect.
Owner:BEIJING CENT FOR PHYSICAL & CHEM ANALYSIS
Amplification primers for identification of olea europaea l. varieties based on SNP loci, screening method and identification method
ActiveCN109554501AClear peak shapeInexpensive testingMicrobiological testing/measurementDNA/RNA fragmentationGenome alignmentScreening method
The invention discloses amplification primers for identification of olea europaea l. varieties based on SNP loci, a screening method, an identification method and 8 pairs of olea europaea l. amplifiedprimers. By restriction endonuclease digestion, clone sequencing and genome comparison analysis of olea europaea l. varieties leaf DNA, high reliability and repeatability single-copy fragments are selected, and the single-copy fragments are selected to design amplification primers; then the amplification primers designed by the selected single-copy fragments are used for amplifying olea europaeal. varieties leaf DNA by PCR, and 8 pairs of single-copy nuclear gene markers with high amplification efficiency and rich variation are screened out. The 8 pairs of olea europaea l. amplified productsare relatively pure and have clear peak shapes and no need of fluorescent primers, and the detection cost is low. The 8 pairs of olea europaea l. Single-copy nuclear gene markers can be used for identifying the olea europaea l. varieties.
Owner:INST OF FORESTRY CHINESE ACAD OF FORESTRY +1
Three newly-separated vibrio fast-growing strains and application thereof
ActiveCN111500484AGrow fastAchieve knockoutBacteriaMicroorganism based processesBiotechnologyAmino acid synthesis
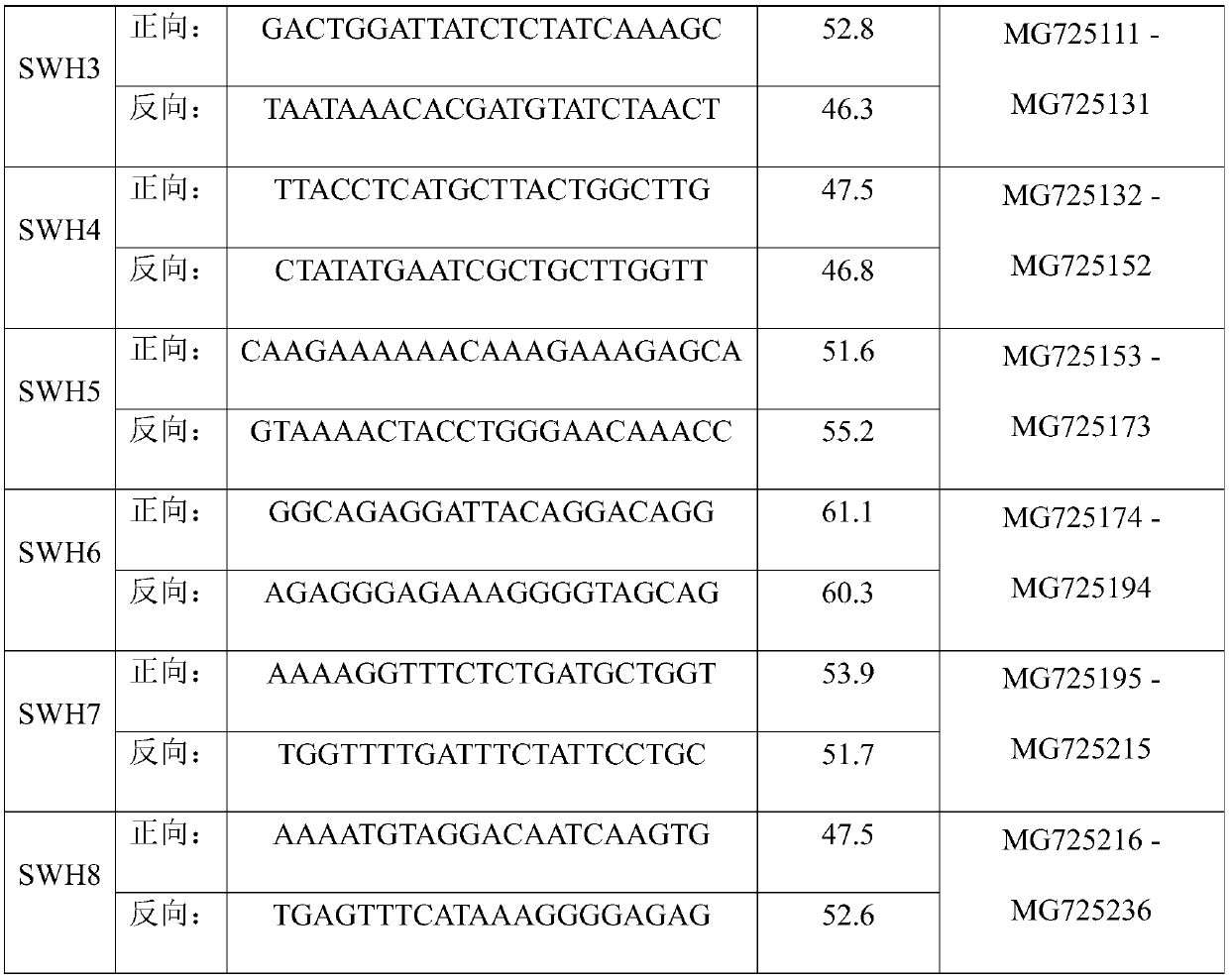
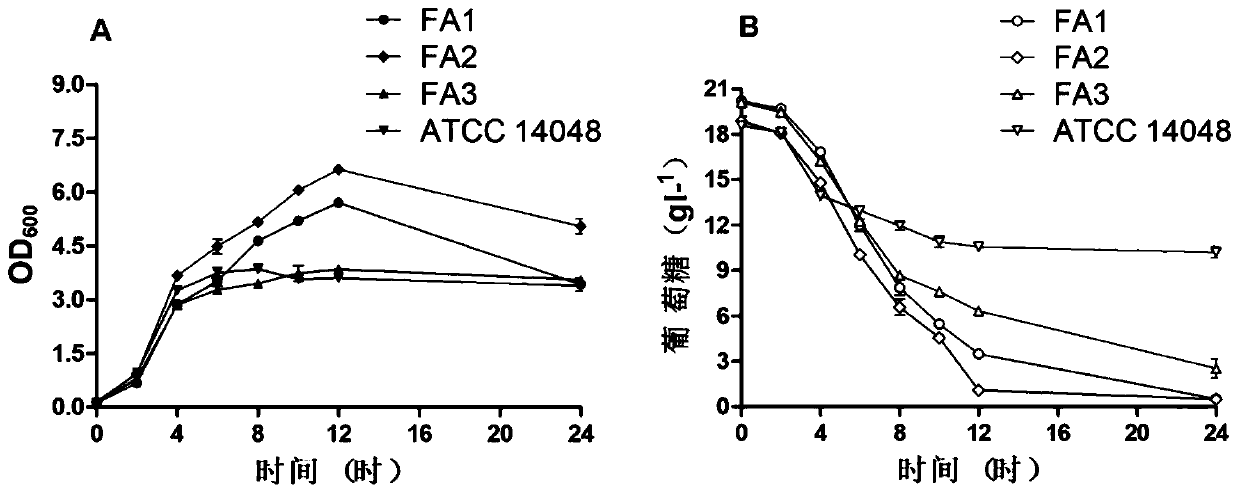
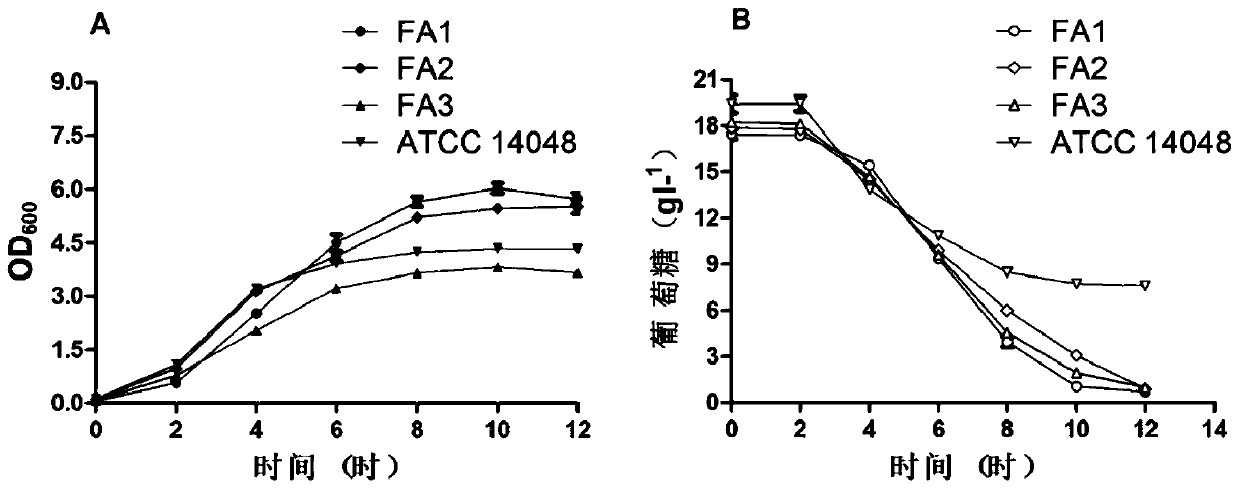
The invention discloses three newly-separated vibrio fast-growing strains and the application of the vibrio fast-growing strains and relates to the field of bioengineering. The three strains of vibrios are respectively named as Vibrio sp. FA1, Vibrio sp. FA2 and Vibrio sp. FA3, and the three strains of vibrio sp. FA2 and Vibrio sp. FA3 are respectively named as Vibrio sp. The strain is preserved in China Center for Type Culture Collection on August 5, 2019, the address is Wuhan University, China, and the preservation numbers are CCTCC NO: M 2019603, CCTCC NO: M 2019604 and CCTCC NO: M 2019605in sequence. Compared with the known Vibrio natriensis ATCC 14048 with the fastest growth speed, the three strains of vibrio natriensis ATCC 14048 have the obvious growth advantages that the vibrio natriensis ATCC 14048 has the highest growth speed; whole genome comparative analysis finds that more DNA replication-related genes, amino acid synthesis-related genes and stress resistance-related genes exist; the three vibrios can quickly grow by adopting various carbon sources under the condition of a chemical synthesis culture medium, and industrial application is facilitated. In addition, electro-transformation operation transformation plasmids and gene knockout prove the genetic operability of the strains, and important foundation and basis are provided for application of the three vibriosin the technical field of industry.
Owner:SHANGHAI JIAO TONG UNIV
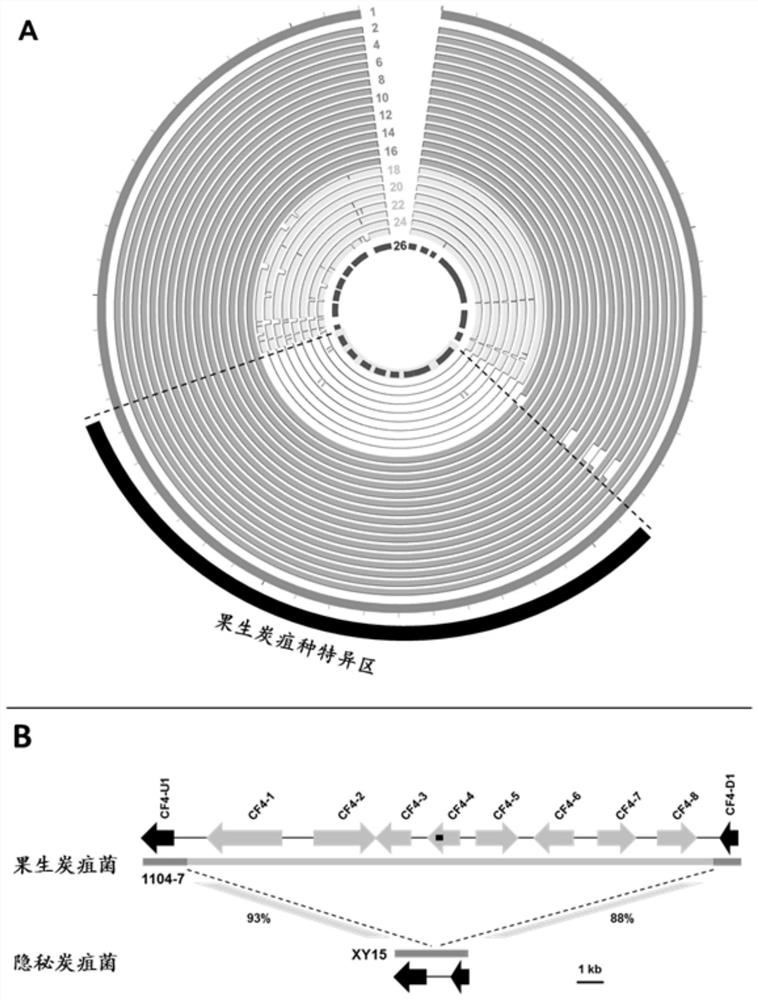
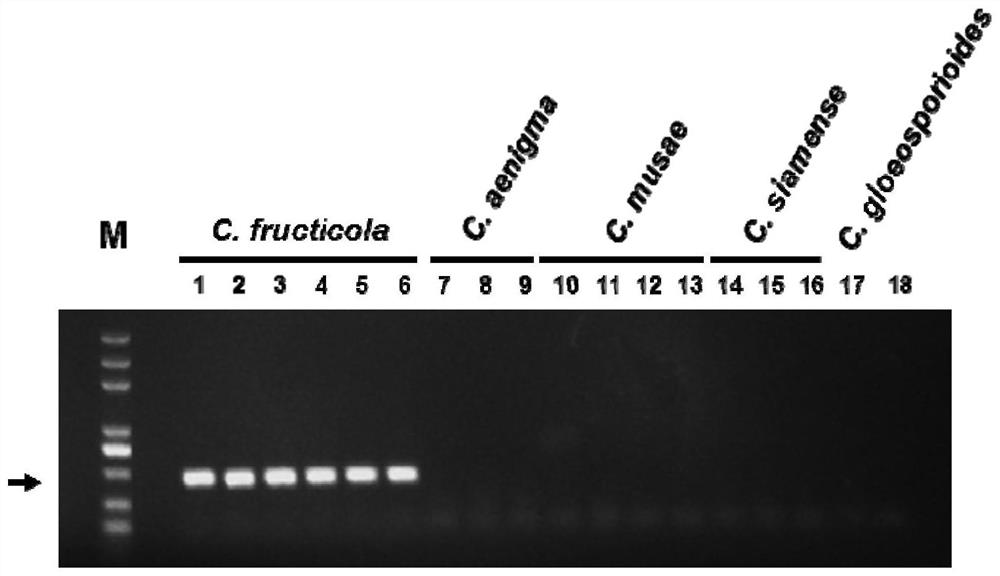
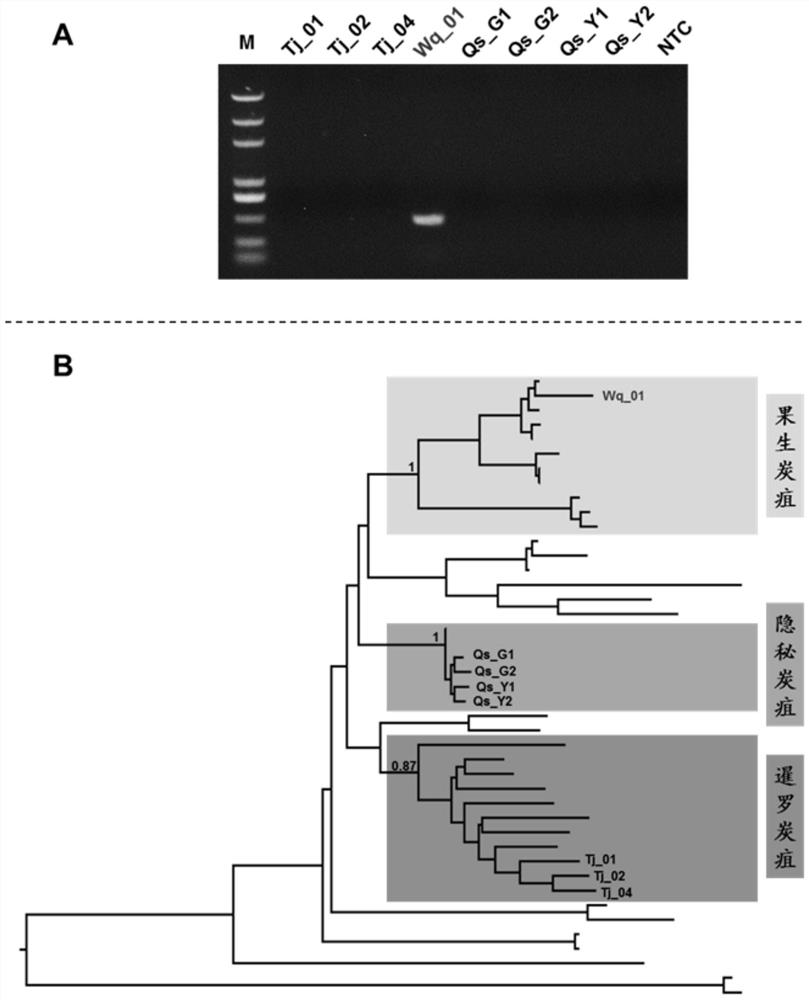
Specific gene sequence of colletotrichum fructicola and application of specific gene sequence
ActiveCN111826459AStrong specificityGood repeatabilityMicrobiological testing/measurementFermentationGenome alignmentGenetics
The invention discloses a specific gene sequence of colletotrichum fructicola and application of the specific gene sequence. According to the invention, the specific gene sequence of the colletotrichum fructicola is obtained based on whole genome comparison of the colletotrichum fructicola and closely related species thereof, and the specific gene sequence is further utilized to design a polymerase chain reaction (PCR) amplification primer to detect the colletotrichum fructicola, and a related kit is developed. The invention can specifically detect the colletotrichum fructicola (C. fructicola)and distinguishes the colletotrichum fructicola from the closely related species including colletotrichum aenigma (C. aenigma), colletotrichum siamense (C. siamense), colletotrichum musae (C. musae)and the like; and the invention has the advantages of being fast, simple, convenient and accurate.
Owner:NORTHWEST A & F UNIV
Genome-scale metabolic network model of fk506-producing strain Streptomyces tsukuba to guide next-level pathway transformation
InactiveCN103279689BIncrease production levelsImprove the experimental effectSpecial data processing applicationsMicroorganismGene cluster
The invention discloses a secondary approach transformation method based on an instruction of an FK506 production bacterial strain wave chain streptomycete genome scale metabolic network model. The model is based on annotation genes and physiology and biochemistry information. By comparing and analyzing the model with a streptomyces coelicolor genome, metabolic genes are found being highly conservative. Metabolic flux analysis is performed on a genome scale metabolic network, and therefore the model predicts a mutation bacterium secondary approach gene cluster transformation strategy for improving a production level. According to the secondary approach transformation method based on the instruction of the FK506 production bacterial strain wave chain streptomycete genome scale metabolic network model, the transformation method utilizes the genome scale metabolic network model to predict special structural genes in an FK506 bacterial strain secondary approach gene cluster, the production level of bacterial strains after transformation is improved by 20 percent to 90 percent, the special structural genes in the gene cluster are augmented to improve production capacity, and large application value is achieved in secondary approach rational transformation of microorganism immunosuppressor production bacterial strains. The high-efficiency and systematic method is provided for optimizing of the bacterial strains.
Owner:TIANJIN UNIV
Genome data storage method and electronic device
ActiveCN107480466BImprove efficiencyInput/output to record carriersBiostatisticsTerm memoryGenomic data
The invention discloses a genomic-data storage method. The genomic-data storage method includes the steps that in the genome comparison process, gene-sequence comparison information is obtained, and gene-sequence statistical information is established; the gene-sequence comparison information is stored in a magnetic disk, and according to comparison positions of the gene-sequence comparison information in a genome, corresponding indexes are stored in a memory, wherein the indexes are storage positions of the gene-sequence comparison information in the magnetic disk; genome statistical information is classified, and first statistical information and second statistical information are obtained; the first statistical information is stored in the memory, wherein the first statistical information is statistical information with the access frequency higher than the preset frequency in the variation detection process; the second statistical information is stored in the magnetic disk, wherein the second statistical information is statistical information which cannot be stored in the memory and / or statistical information with the access frequency lower than the preset frequency in the variation detection process. The invention also discloses an electronic device with the genomic-data storage method.
Owner:UNITED ELECTRONICS
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com