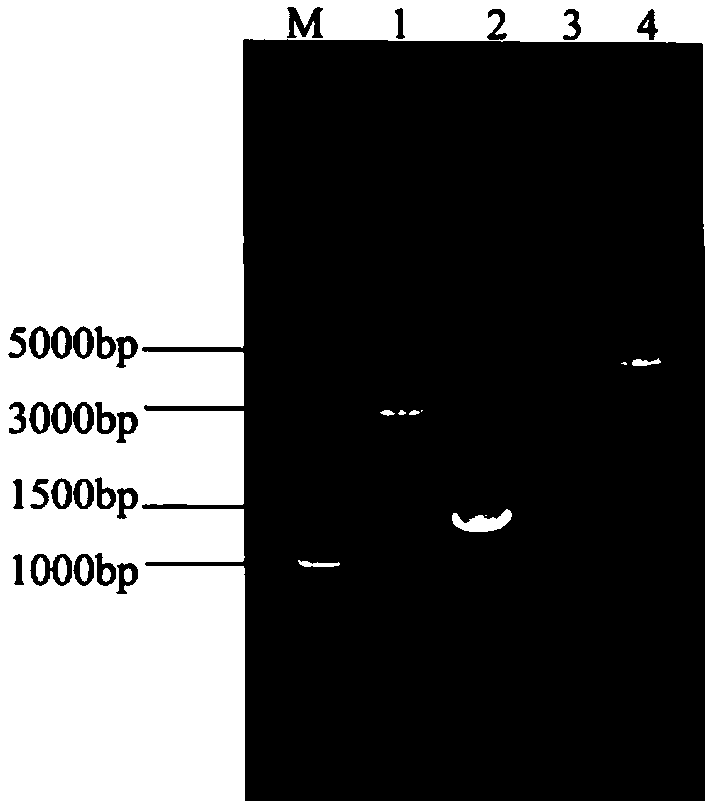
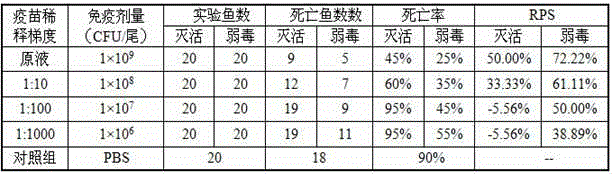
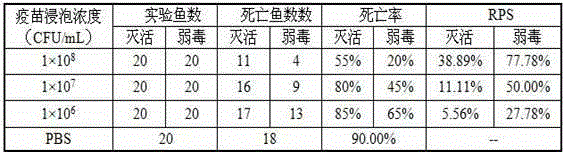
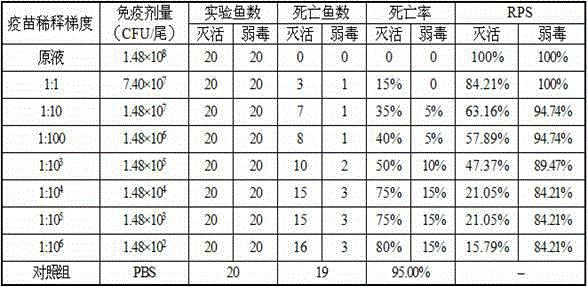
Patents
Literature
249 results about "Streptococcus agalactiae" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Streptococcus agalactiae (also known as group B streptococcus or GBS) is a gram-positive coccus (round bacterium) with a tendency to form chains (as reflected by the genus name Streptococcus). It is a beta-hemolytic, catalase-negative, and facultative anaerobe. In general, GBS is a harmless commensal bacterium being part of the human microbiota colonizing the gastrointestinal and genitourinary tract of up to 30% of healthy human adults (asymptomatic carriers). Nevertheless, GBS can cause severe invasive infections.
Probe arrays for detecting multiple strains of different species
InactiveUS20060160121A1Growth inhibitionReduced virulenceBioreactor/fermenter combinationsBiological substance pretreatmentsStreptococcus pyogenesTO-18
The present invention provides probe arrays and methods of using the same for concurrent and discriminable detection of multiple strains of different species. In one aspect, the probe arrays of the present invention are nucleic acid arrays comprising (1) a first group of probes, each of which is specific to a different respective strain of a first species; and (2) a second group of probes, each of which is specific to a different respective strain of a second species. In many embodiments, the nucleic acid arrays of the present invention further include a third group of probes, each of which is specific to a different strain of a third species. In one example, a nucleic acid array of the present invention includes probes for sequences selected from SEQ ID NOs: 1 to 18,598, and can discriminably detect different strains of Streptococcus pyogenes, Streptococcus agalactiae and Staphylococcus epidermidis.
Owner:WYETH LLC
Immunogenic compositions for gram positive bacteria such as streptococcus agalactiae
InactiveUS20060165716A1InhibitionEasy to eliminateAntibacterial agentsBacterial antigen ingredientsBacteroidesVirulent characteristics
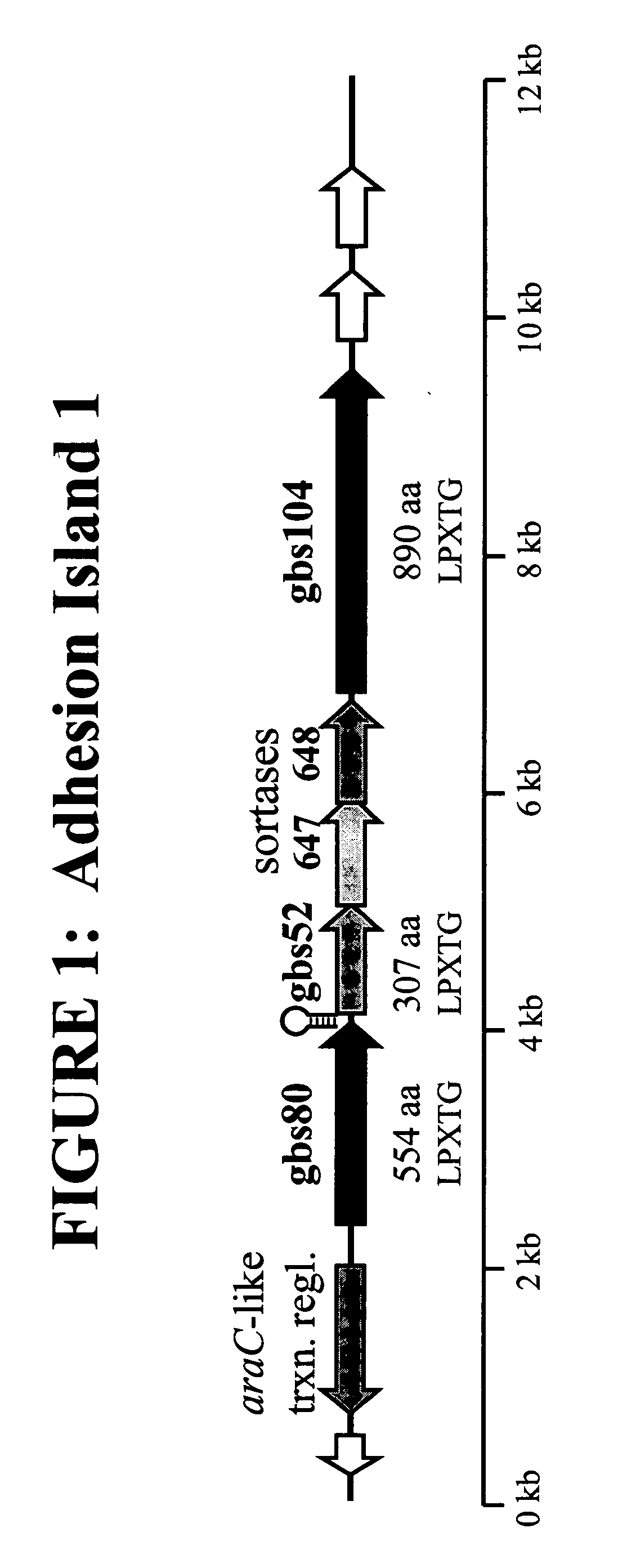
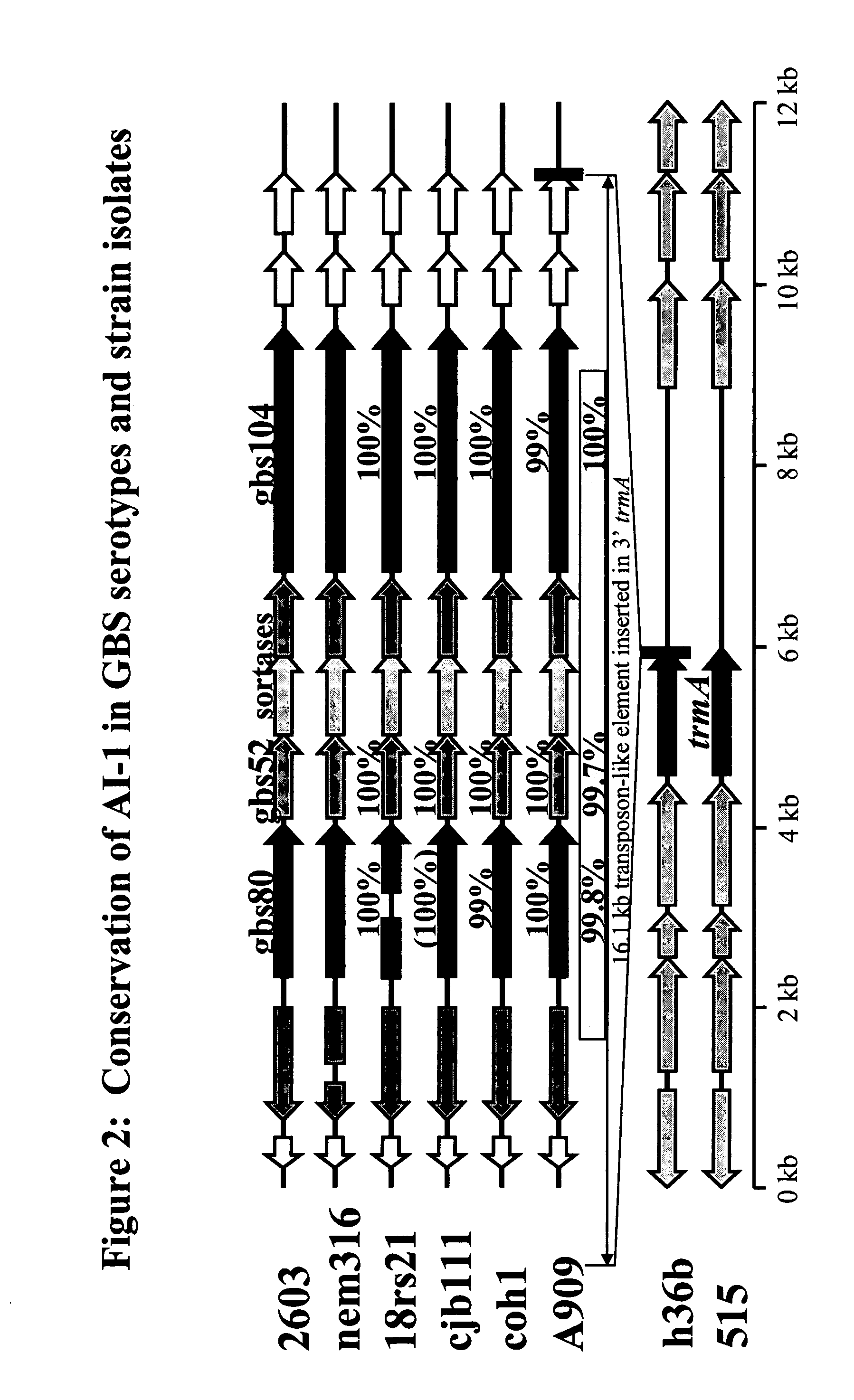
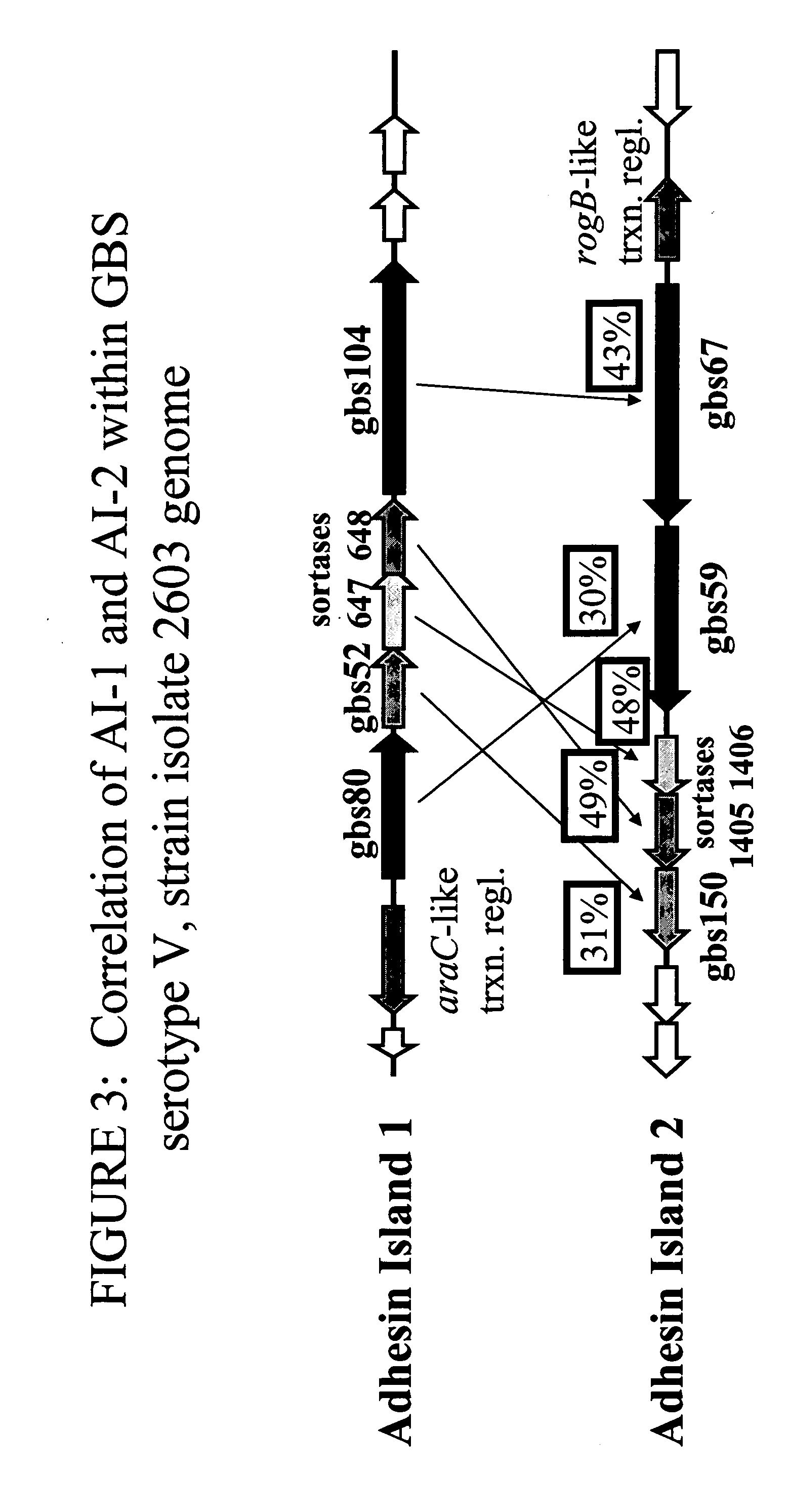
The invention relates to the identification of a new adhesin islands within the genomes of several Group A and Group B Streptococcus serotypes and isolates. The adhesin islands are thought to encode surface proteins which are important in the bacteria's virulence. Thus, the adhesin island proteins of the invention may be used in immunogenic compositions for prophylactic or therapeutic immunization against GAS or GBS infection. For example, the invention may include an immunogenic composition comprising one or more of the discovered adhesin island proteins.
Owner:NOVARTIS VACCINES & DIAGNOSTICS INC
Nucleic acids and proteins from streptococcus groups A & B
InactiveUS20060210580A1Antibacterial agentsSenses disorderStreptococcus pyogenesStreptococcus agalactiae
The invention provides proteins from group B streptococcus (Streptococcus agalactiae) and group A streptococcus (Streptococcus pyogenes), including amino acid sequences and the corresponding nucleotide sequences. Data are given to show that the proteins are useful antigens for vaccines, immunogenic compositions, and / or diagnostics. The proteins are also targets for antibiotics.
Owner:GLAXOSMITHKLINE BIOLOGICALS SA
Nucleic acids and proteins from streptococcus groups A & B
InactiveUS20060210579A1Antibacterial agentsSenses disorderStreptococcus pyogenesStreptococcus agalactiae
The invention provides proteins from group B streptococcus (Streptococcus agalactiae) and group A streptococcus (Streptococcus pyogenes), including amino acid sequences and the corresponding nucleotide sequences. Data are given to show that the proteins are useful antigens for vaccines, immunogenic compositions, and / or diagnostics. The proteins are also targets for antibiotics.
Owner:NOVARTIS VACCINES & DIAGNOSTICS INC
Nucleic acids and proteins from streptococcus groups a & b
InactiveUS20060275315A1Easy to insertEfficient HarvestingAntibacterial agentsBacteriaStreptococcus pyogenesStreptococcus agalactiae
The invention provides proteins from group B streptococcus (Streptococcus agalactiae) and group A streptococcus (Streptococcus pyogenes), including amino acid sequences and the corresponding nucleotide sequences.
Owner:CHIRON CORP +1
Nucleic acids and proteins from streptococcus groups A & B
InactiveUS20060210582A1Antibacterial agentsSenses disorderStreptococcus pyogenesStreptococcus agalactiae
The invention provides proteins from group B streptococcus (Streptococcus agalactiae) and group A streptococcus (Streptococcus pyogenes), including amino acid sequences and the corresponding nucleotide sequences. Data are given to show that the proteins are useful antigens for vaccines, immunogenic compositions, and / or diagnostics. The proteins are also targets for antibiotics.
Owner:GLAXOSMITHKLINE BIOLOGICALS SA
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20040185478A1Reduce usageDetermine rapidly the bacterial resistance to antibioticsMicrobiological testing/measurementFermentationBacteroidesNeisseria meningitidis
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6'-lla, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6')-aph(2''), aad(6'), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:GENEOHM SCI CANADA
Nucleic acids and proteins from streptococcus groups A & B
The invention provides proteins from group B streptococcus (Streptococcus agalactiae) and group A streptococcus (Streptococcus pyogenes), including amino acid sequences and the corresponding nucleotide sequences. Data are given to show that the proteins are useful antigens for vaccines, immunogenic compositions, and / or diagnostics. The proteins are also targets for antibiotics.
Owner:GLAXOSMITHKLINE BIOLOGICALS SA
Immunogenic Compositions for Streptococcus Agalactiae
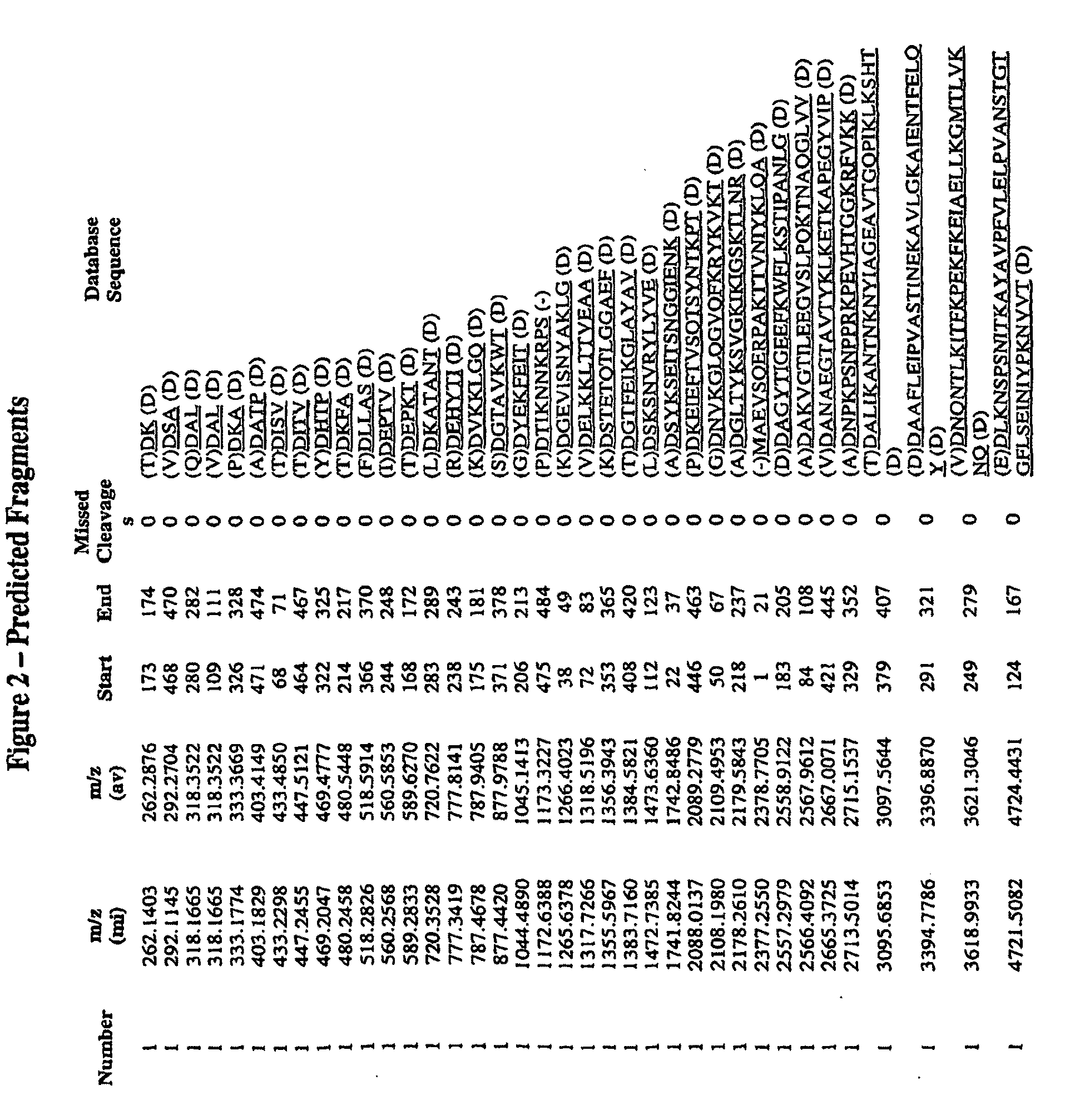
This application relates to Group B Streptococcus (“GBS”) vaccines comprising combinations of GBS polypeptide antigens where the polypeptides contribute to the immunological response in a recipient. Preferably, the compositions of the invention comprise a combination of two or more GBS antigens, wherein said combination includes GBS 80 or a fragment thereof. In one embodiment, the combination may consist of two to thirteen GBS antigens selected from an antigen group consisting of GBS 80, GBS 91, GBS 104, GBS 184, GBS 276, GBS 305, GBS 322, GBS 330, GBS 338, GBS 361, GBS 404, GBS 690, and GBS 691.
Owner:CHIRON CORP
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20060263810A1Reduce usageDetermine rapidly the bacterial resistance to antibioticsSugar derivativesMicrobiological testing/measurementNeisseria meningitidisListeria monocytogenes
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6′-IIa, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6′)-aph(2″), aad(6), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:GENEOHM SCI CANADA
Nucleic acids and proteins from streptococcus groups A & B
InactiveUS20060210581A1Antibacterial agentsSenses disorderStreptococcus pyogenesStreptococcus agalactiae
The invention provides proteins from group B streptococcus (Streptococcus agalactiae) and group A streptococcus (Streptococcus pyogenes), including amino acid sequences and the corresponding nucleotide sequences. Data are given to show that the proteins are useful antigens for vaccines, immunogenic compositions, and / or diagnostics. The proteins are also targets for antibiotics.
Owner:GLAXOSMITHKLINE BIOLOGICALS SA
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20030049636A1Low costRapid positioningMicrobiological testing/measurementFermentationBacteroidesNeisseria meningitidis
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6'-lla, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6')-aph(2''), aad(6'), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:BERGERON MICHEL G +3
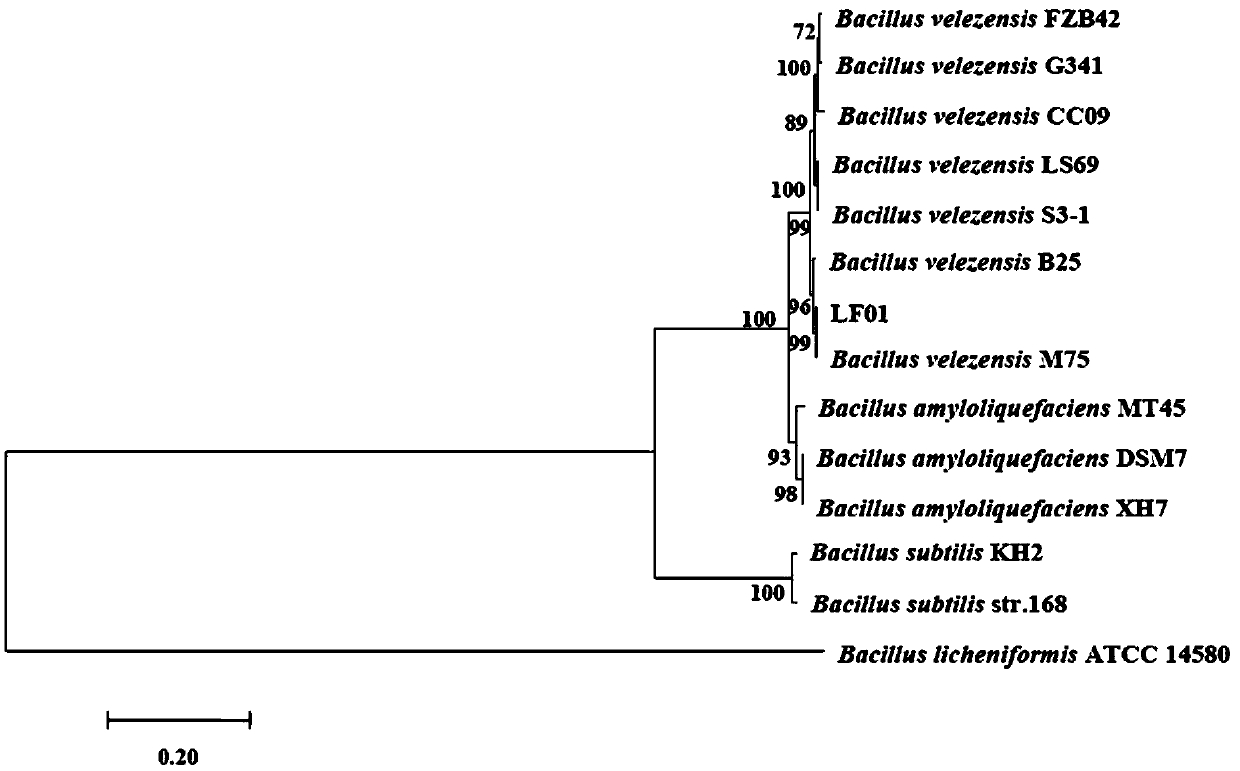
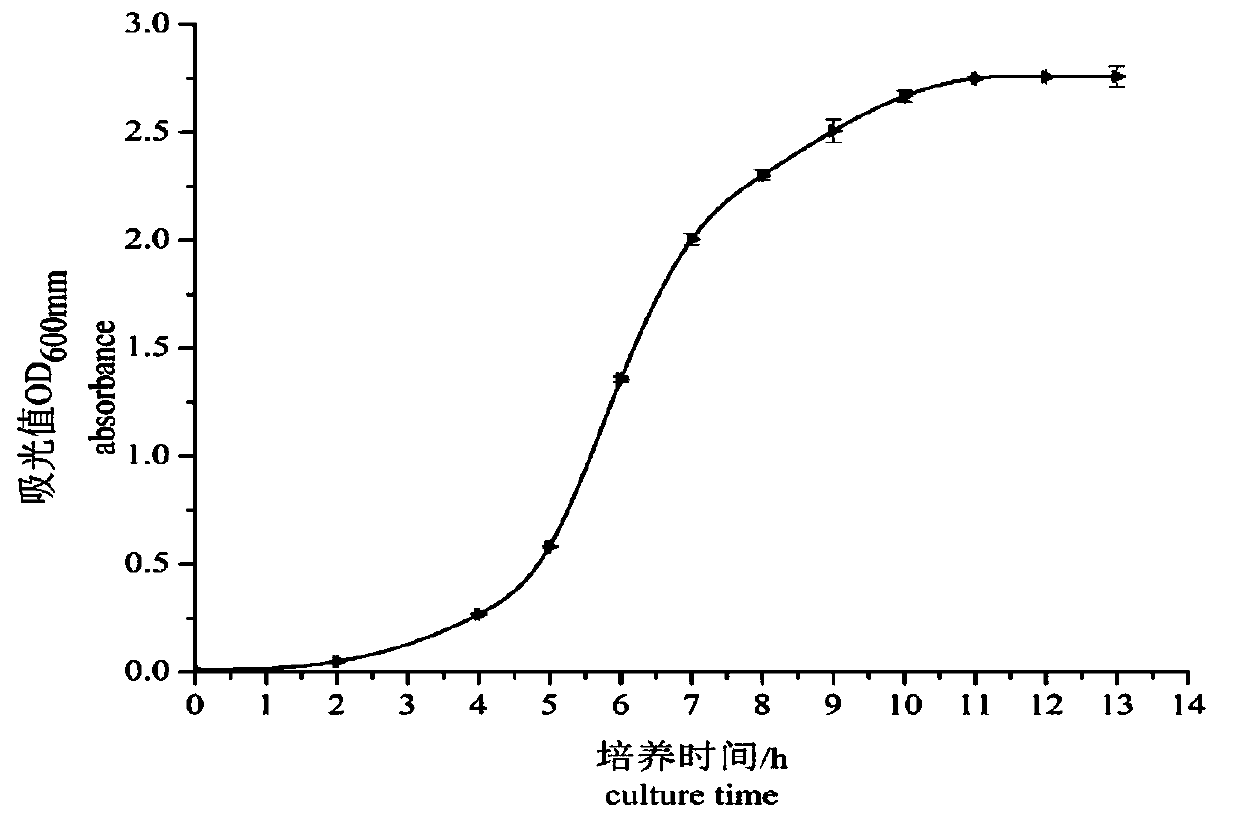
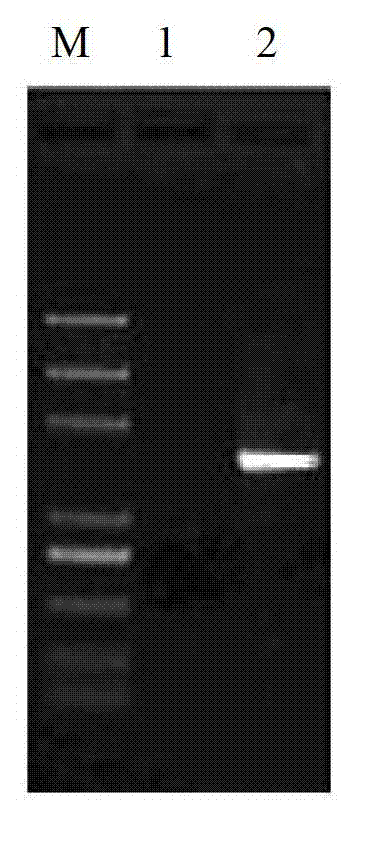
Bacillus velezensis and application of bacillus velezensis as aquatic pathogenic bacteria inhibitor
ActiveCN108676756AEnhance immune functionStrong antibacterial functionAntibacterial agentsBacteriaBacterial strainAquaculture
The invention provides bacillus velezensis and application of the bacillus velezensis as an aquatic pathogenic bacteria inhibitor. The bacillus velezensis is separated from the intestinal canal of healthy tilapia, and the preservation number of the bacterial strain is GDMCC No. 60344. The bacillus velezensis has a broad-spectrum antagonistic aquatic common pathogenic bacteria function, and has theadvantages that the growth of the pathogenic bacteria, such as streptococcus agalactiae, streptococcus iniae,nocardia seriolea, aeromonas hydrophila, aeromonas schubertii and edwardsiella tarda, canbe effectively inhibited, the use is safe, no residue is generated, reducing the usage amount and the residual amount of chemical drugs in aquaculture is facilitated, the production cost is reduced, and the practicality is strong.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
Vaccine used for preventing tilapia streptococcal disease
ActiveCN102847146AHigh protection rateEasy to prepareAntibacterial agentsBacterial antigen ingredientsStreptococcus agalactiaeTilapia
The invention discloses a vaccine used for preventing tilapia streptococcal disease. The vaccine is prepared by using streptococcus agalactiae and pharmaceutically acceptable excipients or carriers. The invention discloses a purpose of the vaccine in preparing medicines used for preventing tilapia streptococcal disease, and a combinatively used medicine. The invention also discloses a novel streptococcus agalactiae strain. The vaccine provided by the invention can effectively stimulate tilapia to produce immunity resisting streptococcal disease. The vaccine has a high protection rate in preventing tilapia streptococcal disease, and has high practical value and good economic benefit.
Owner:TONGWEI
Group b streptococcus polysaccharide-protein conjugates, methods for producing conjugates, immunogenic compositions comprising conjugates, and uses thereof
ActiveUS20160324950A1Antibacterial agentsBacterial antigen ingredientsPassive ImmunizationsAntiendomysial antibodies
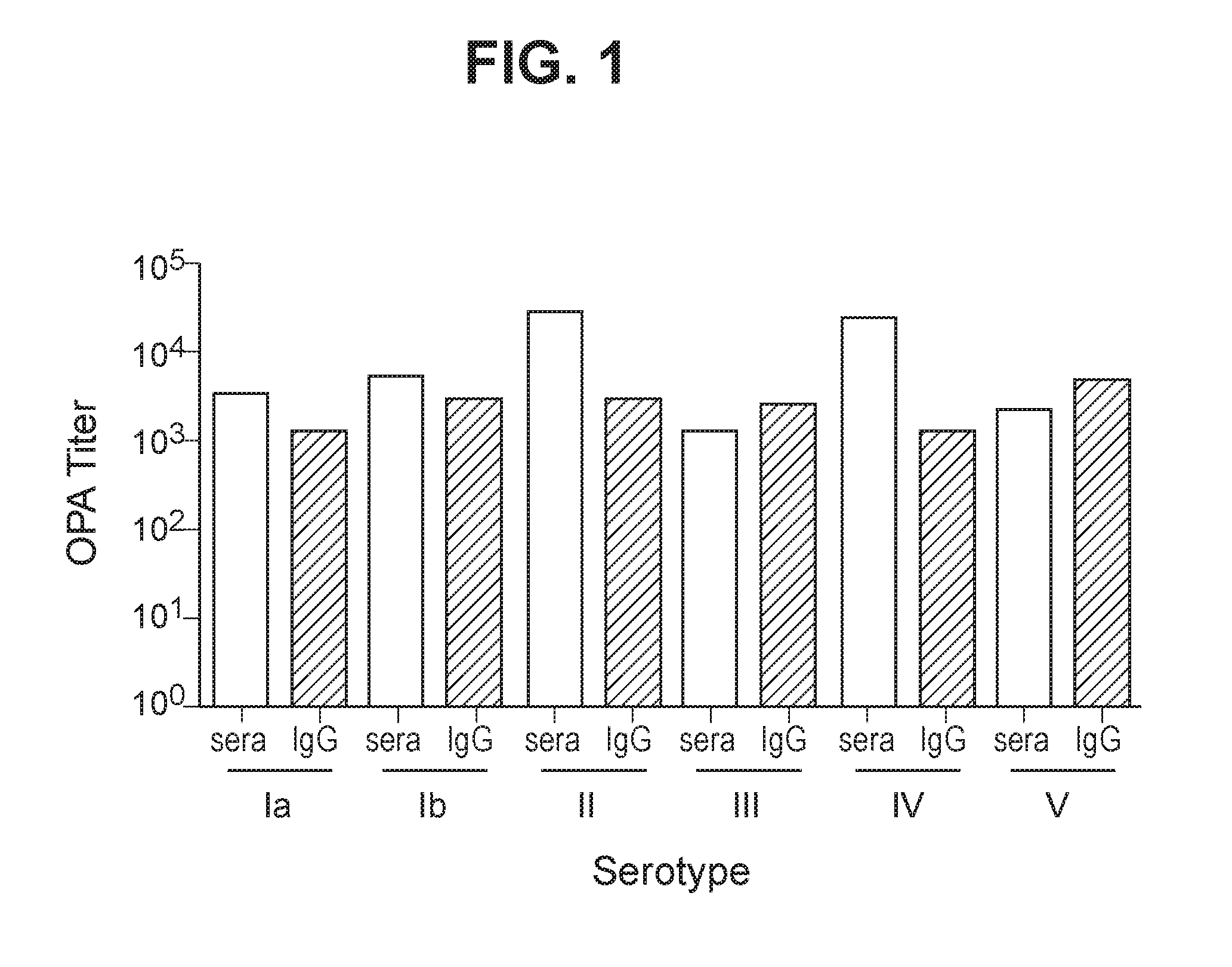
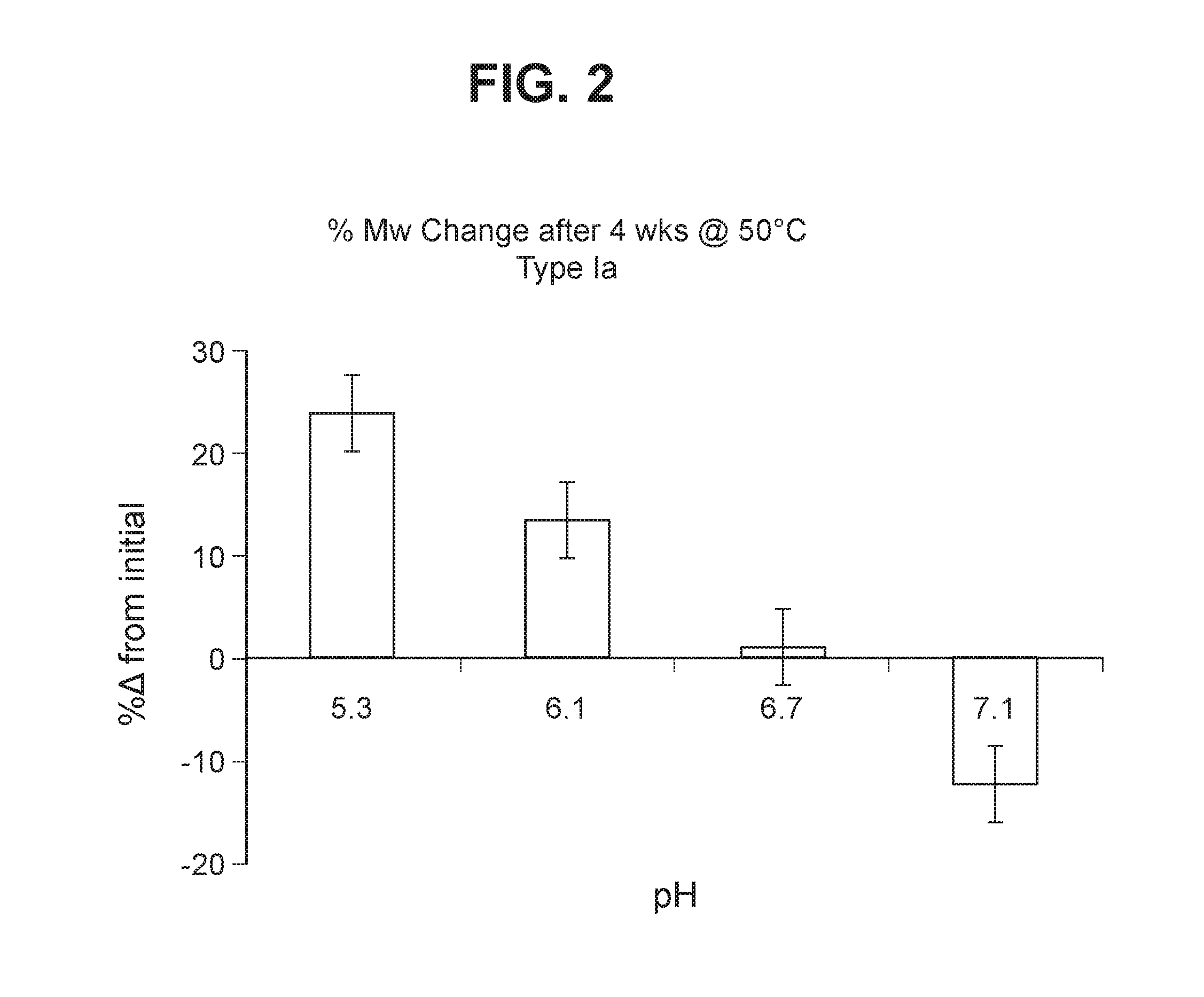
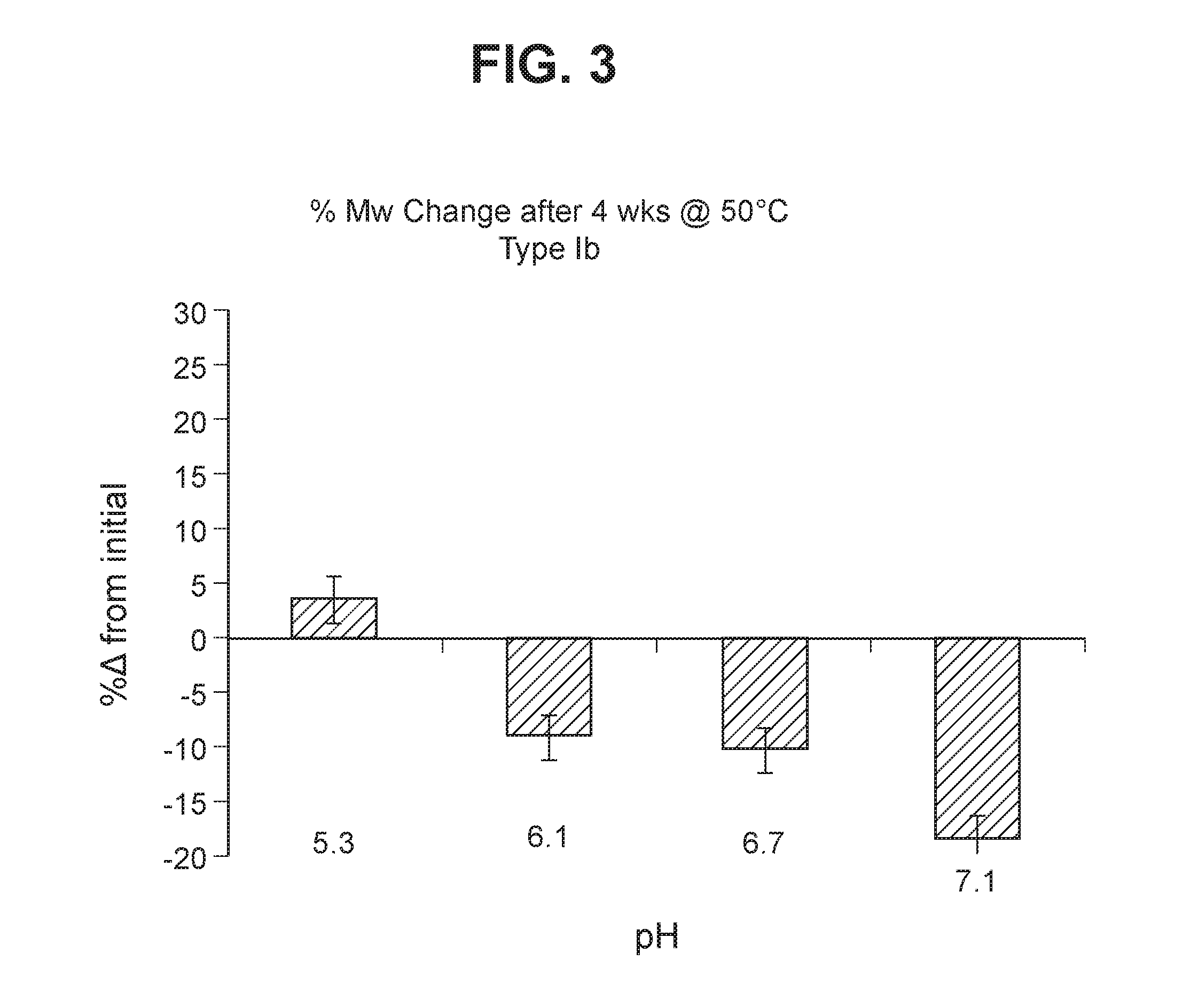
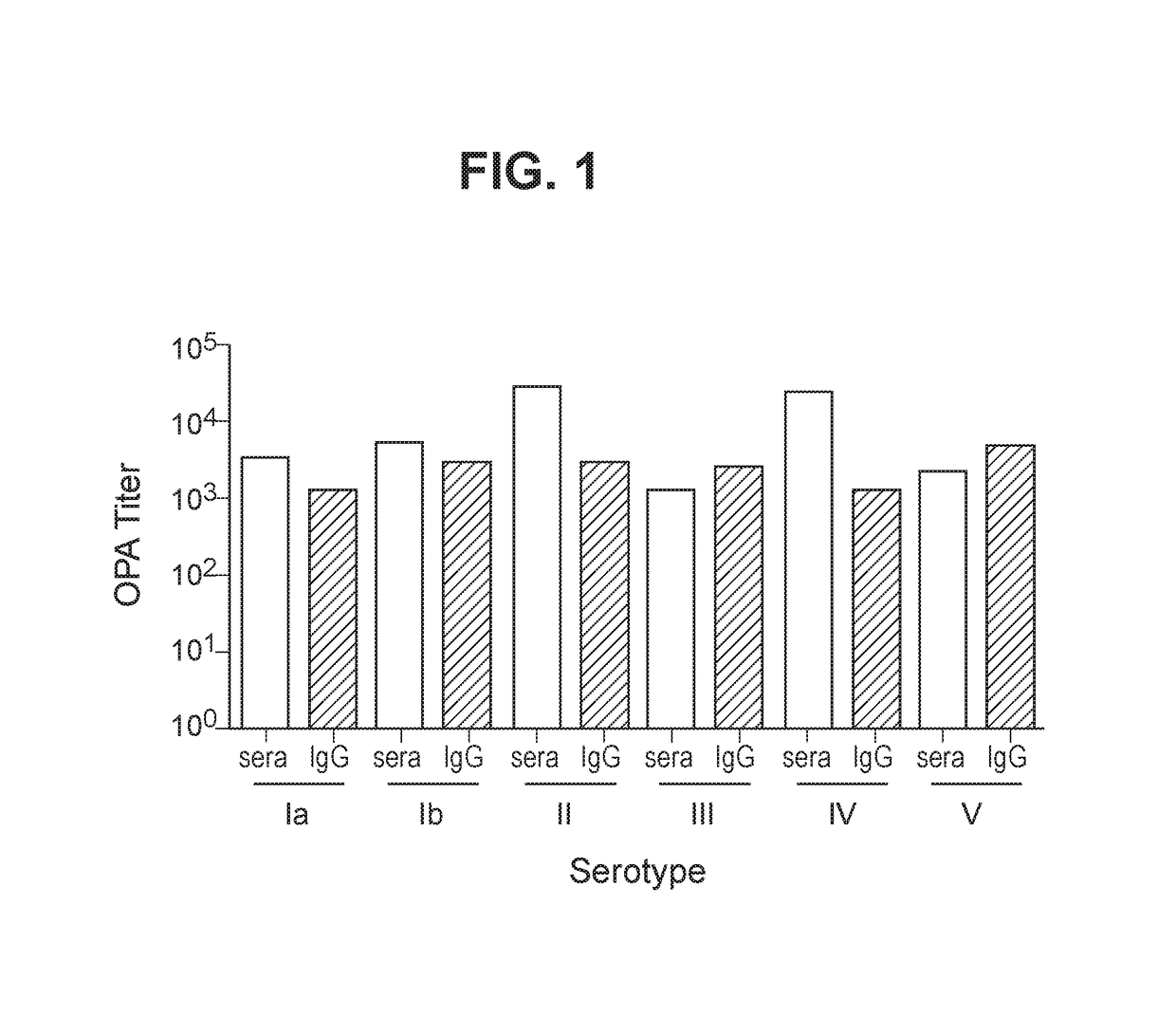
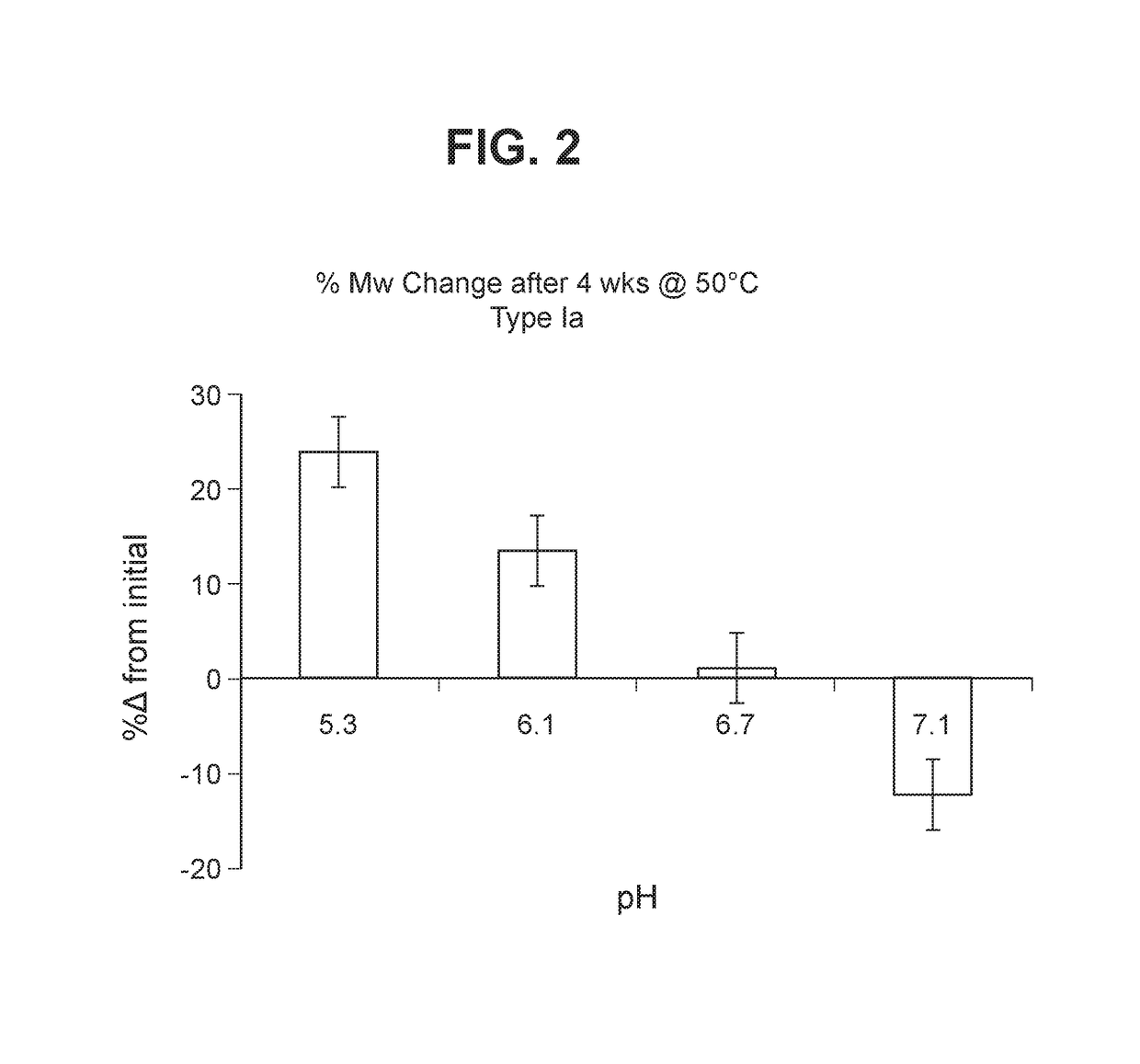
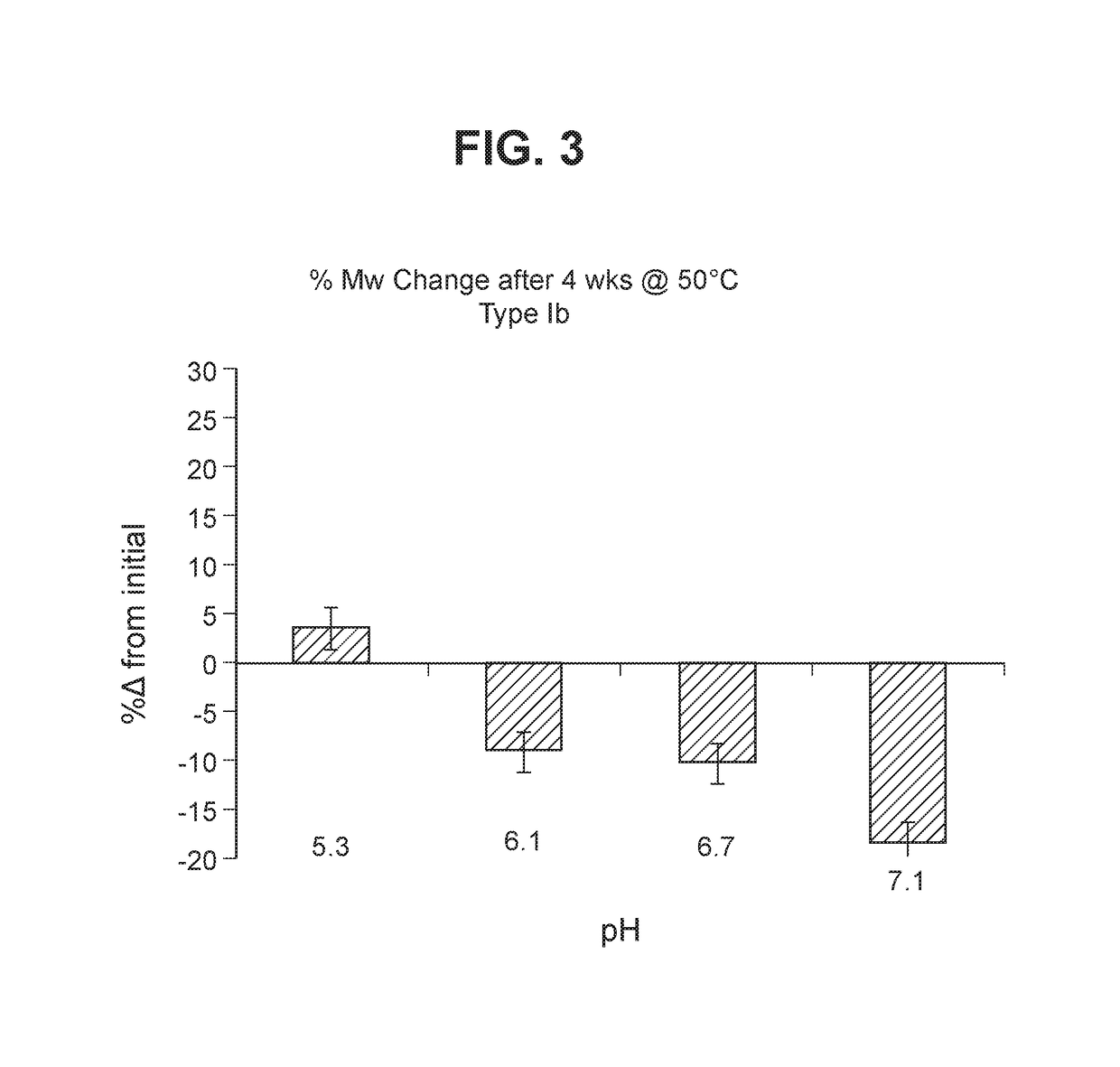
The invention relates to immunogenic polysaccharide-protein conjugates comprising a capsular polysaccharide (CP) from Streptococcus agalactiae, commonly referred to as group B streptococcus (GBS), and a carrier protein, wherein the CP is selected from the group consisting of serotypes Ia, Ib, II, III, IV, V, VI, VII, VIII, and IX, and wherein the CP has a sialic acid level of greater than about 60%. The invention also relates to methods of making the conjugates and immunogenic compositions comprising the conjugates. The invention also relates to immunogenic compositions comprising polysaccharide-protein conjugates, wherein the conjugates comprise a CP from GBS serotype IV and at least one additional serotype. The invention further relates to methods for inducing an immune response in subjects against GBS and / or for reducing or preventing invasive GBS disease in subjects using the compositions disclosed herein. The resulting antibodies can be used to treat or prevent GBS infection via passive immunotherapy.
Owner:PFIZER INC
Oral vaccine of streptococcus agalactiae and preparation method thereof
ActiveCN103834669AAntibacterial agentsBacterial antigen ingredientsStreptococcus agalactiaeMicrobiology
The invention discloses a nucleotide sequence as shown in SEQ ID No.: 1, and a recombinant vector and recombinant bacteria containing the nucleotide sequence. The invention also discloses oral vaccine of streptococcus agalactiae. The oral vaccine of streptococcus agalactiae is characterized by being grafted and immunized by an oral manner and has the advantages of being high in protection rate for tilapia mossambica, convenient to administrate, simple in operation, small in immunological stress reaction, safe, and high in application prospect.
Owner:TONGWEI
Multiplex PCR kit for detecting bacterial meningitis pathogens
ActiveCN105734164AReduce usageReduce testing costsMicrobiological testing/measurementDNA/RNA fragmentationBacteroidesStaphylococcus aureus
The invention discloses a multiplex PCR kit for detecting bacterial meningitis pathogens. Six types of bacterial meningitis pathogens, namely streptococcus pneumoniae (SP), streptococcus agalactiae (GBS), haemophilus influenzae (HI), listeria monocytogenes (LM), staphylococcus aureus (SA) and neisseria meningitidis (NM) can be rapidly detected. The invention discloses a 'public primer'-mediated multiplex PCR method. The method concretely comprises the steps of constructing a plasmid template, screening a public primer, designing a multi-primer and carrying out multiplex PCR system detection; a two-step annealing temperature method is combined, so that specificity, sensibility and detection throughput of a multiplex PCR system are improved; the multiplex PCR method has a certain containment capability, detection targets can be continuously added, and the detection throughput of multiplex PCR can be beneficially improved. The multiplex PCR kit disclosed by the invention can be used for effectively detecting six types of common bacteria, has the advantages of high sensitivity, economy, high speed and simple operation and is beneficial to application and clinical diagnosis.
Owner:上海捷诺生物科技股份有限公司
Group B Streptococcus polysaccharide-protein conjugates, methods for producing conjugates, immunogenic compositions comprising conjugates, and uses thereof
The invention relates to immunogenic polysaccharide-protein conjugates comprising a capsular polysaccharide (CP) from Streptococcus agalactiae, commonly referred to as group B streptococcus (GBS), and a carrier protein, wherein the CP is selected from the group consisting of serotypes Ia, Ib, II, III, IV, V, VI, VII, VIII, and IX, and wherein the CP has a sialic acid level of greater than about 60%. The invention also relates to methods of making the conjugates and immunogenic compositions comprising the conjugates. The invention also relates to immunogenic compositions comprising polysaccharide-protein conjugates, wherein the conjugates comprise a CP from GBS serotype IV and at least one additional serotype. The invention further relates to methods for inducing an immune response in subjects against GBS and / or for reducing or preventing invasive GBS disease in subjects using the compositions disclosed herein. The resulting antibodies can be used to treat or prevent GBS infection via passive immunotherapy.
Owner:PFIZER INC
Conjugation of streptococcal capsular saccharides
ActiveUS8513392B2Avoid disruptionImproving immunogenicityAntibacterial agentsImmunoglobulinsStreptococcus agalactiaeLactose oxidase
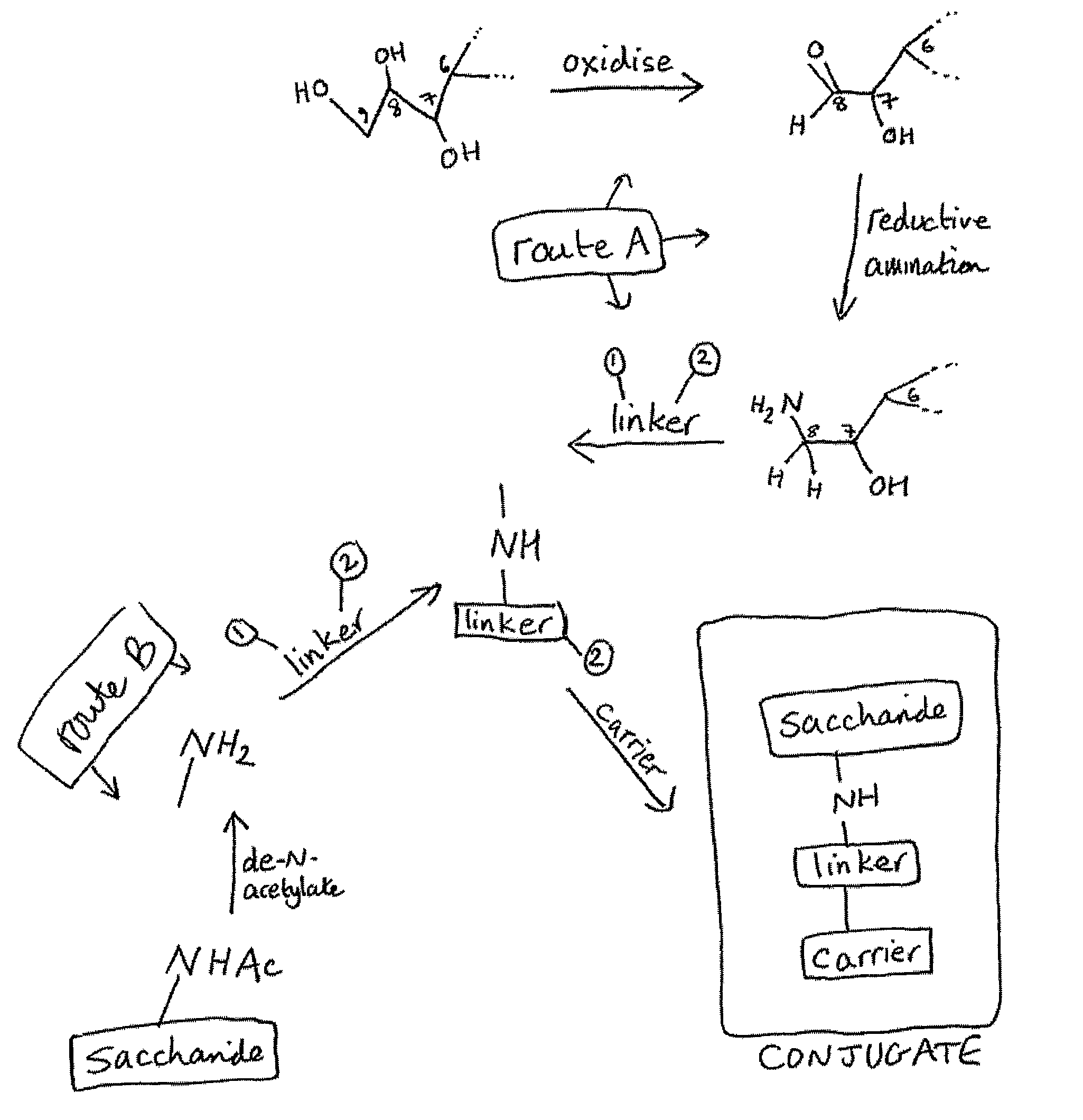
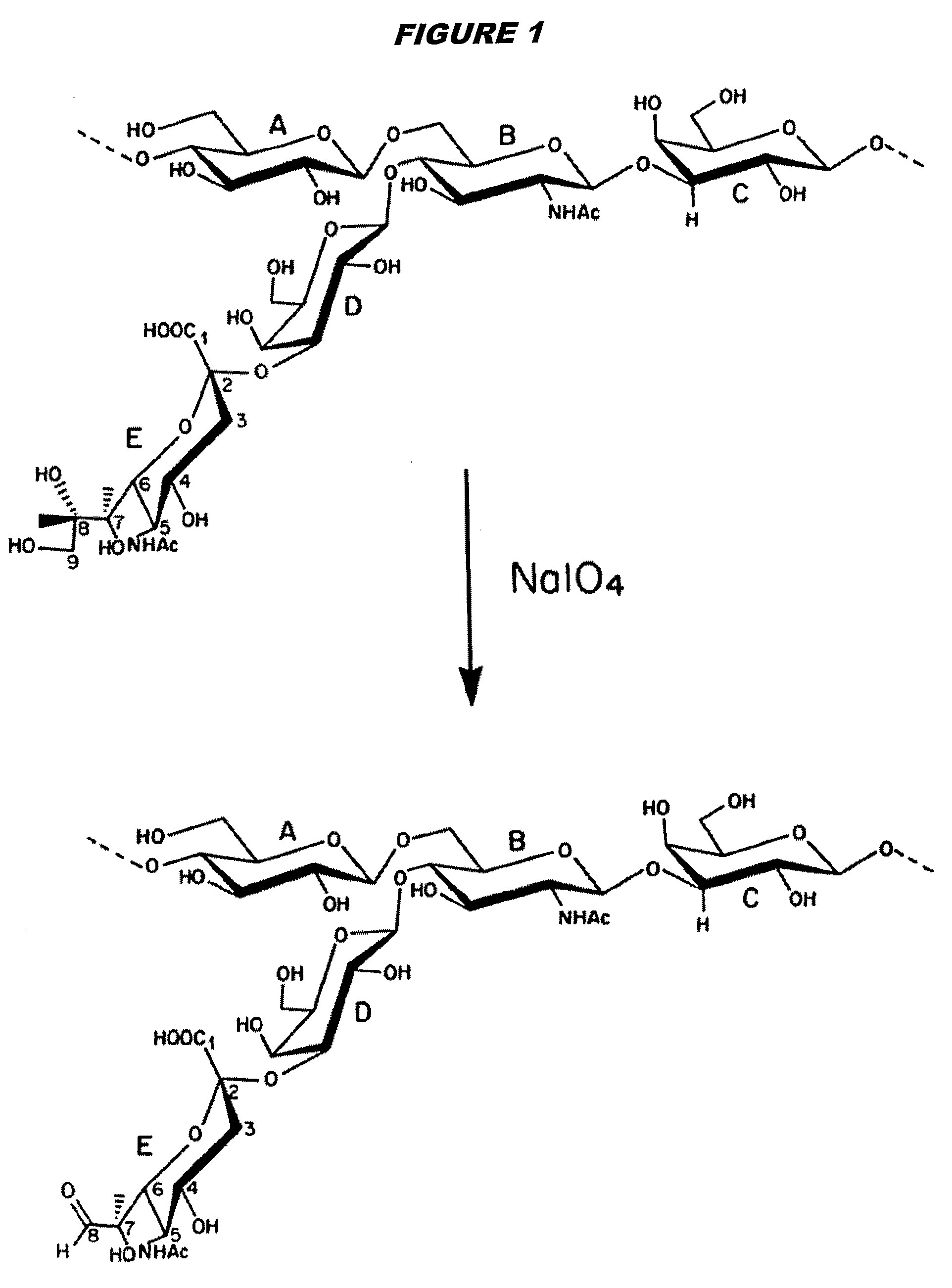
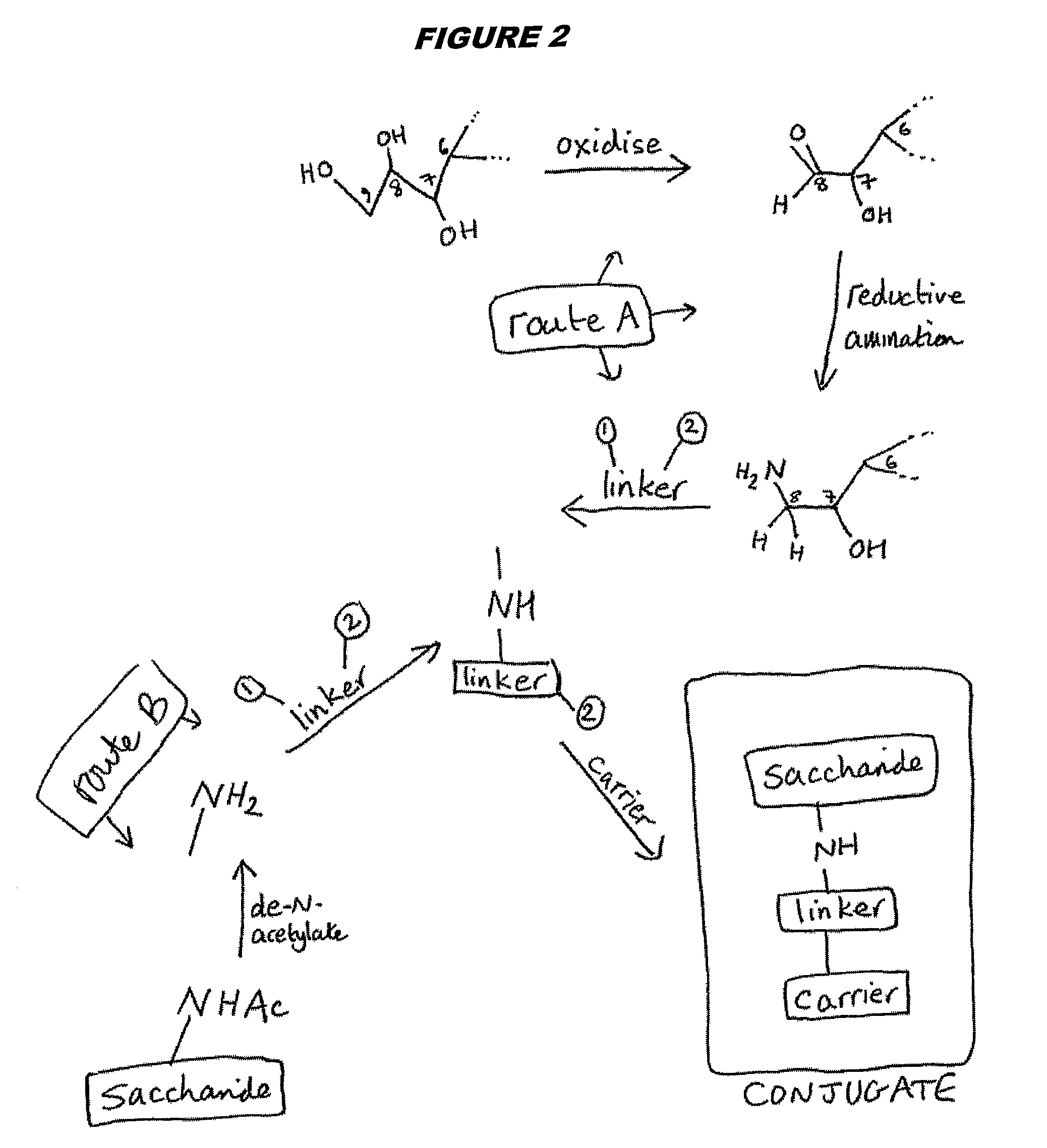
Three conjugation methods for use with the capsular saccharide of Streptococcus agalactiae. In the first method, reductive animation of oxidized sialic acid residue side chains is used, but the aldehyde groups are first aminated, and then the amine is coupled to a carrier via a linker. In the second method, sialic acid residues and / or N-acetyl-glucosamine residues are de-N-acetylated to give amine groups, and the amine groups are coupled to a carrier protein via a linker. In the third method, linkage is via galactose residues in the capsular saccharide rather than sialic acid residues, which can conveniently be achieved using galactose oxidase.
Owner:GLAXOSMITHKLINE BIOLOGICALS SA
Immunogenic compositions for streptococcus agalactiae
InactiveUS20100015168A1Generate immune responseAntibacterial agentsPeptide/protein ingredientsEpitopeStreptococcus agalactiae
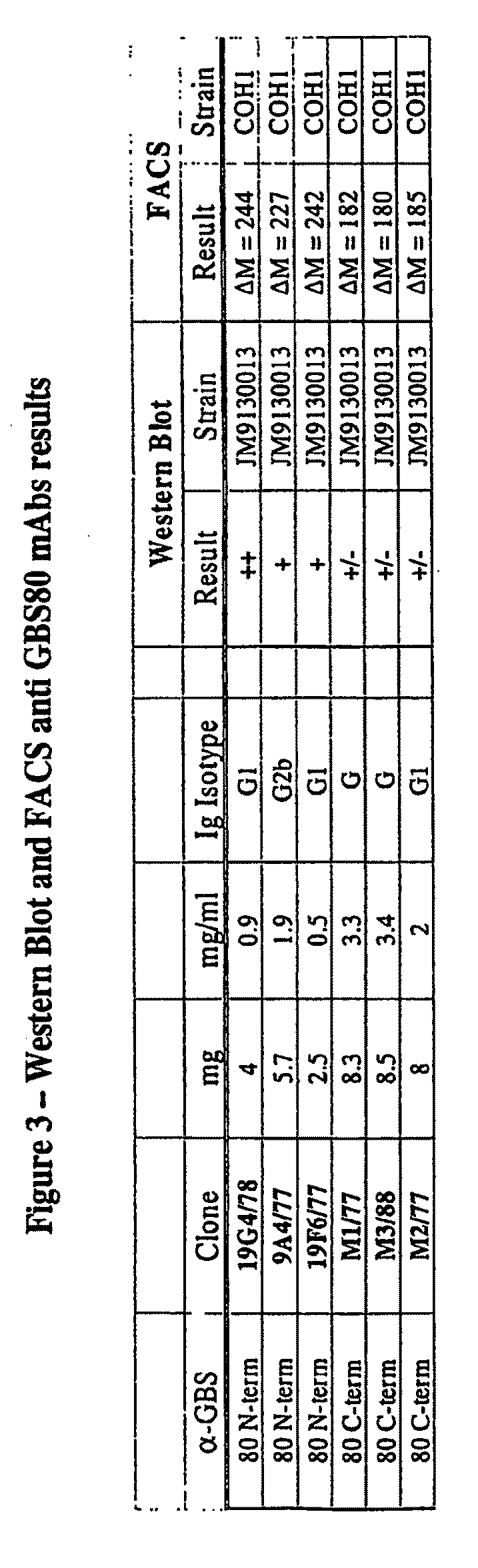
The invention relates to immunogenic polypeptides derived from epitopes in a Streptococcus agalactiae (“GBS”) protein GBS 80 and their use as prophylactic, diagnostic and therapeutic compositions. The invention also provides nucleic acids encoding the immunogenic polypeptides. Also provided are vectors useful for making such immunogenic polypeptides and host cells transformed with such vectors. In particular, the invention relates to a group immunogenic polypeptides derived from GBS 80. The compositions may include one or more of the immunogenic polypeptides either alone or with other antigenic components. For example, the immunogenic polypeptides may be combined with other GBS antigens to provide therapeutic compositions with broader range. In addition, the immunogenic polypeptides may also include flanking portions of the GBS 80 protein.
Owner:NOVARTIS AG
Method and kit for detecting streptococcus agalactiae
ActiveCN102242202AAccurate detectionEfficient detectionMicrobiological testing/measurementMicroorganism based processesDiseaseStreptococcus agalactiae
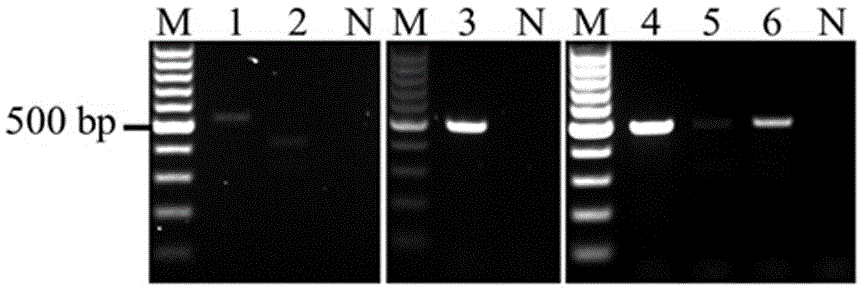
The invention discloses a method and a kit for detecting pathogenic germs of fish, and relates to detection technology for streptococcus agalactiae. The invention provides a kit for detecting detecting pathogenic germs of fish. The kit comprises a primer pair 1 with sequences as represented by SEQ ID NO.1-2 for amplifying genes from streptococcus agalactiae and / or a primer pair 2 with sequences as represented by SEQ ID NO. 3-4 for amplifying genes from streptococcus agalactiae. The invention also provides a method for detecting detecting pathogenic germs of fish with the above-mentioned kit. The detection kit and method provided in the invention can accurately and effectively detect streptococcus agalactiae, have the characteristics of high sensitivity, strong specificity, short time and rapid detection, are applicable to rapid detection, prediction and prevention of fish diseases, and can increase economic benefits.
Owner:TONGWEI
Streptococcus agalactiae loop-mediated isothermal amplification kit and application thereof
InactiveCN104726567AEasy way to judge the resultNo pollutionMicrobiological testing/measurementStreptococcus agalactiaeMicrobiology
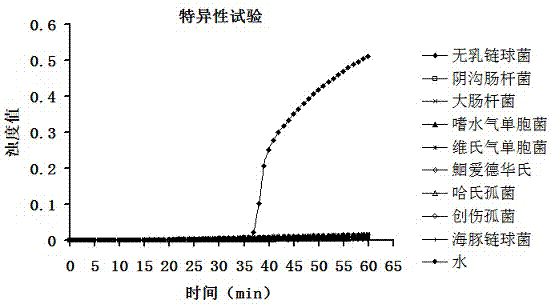
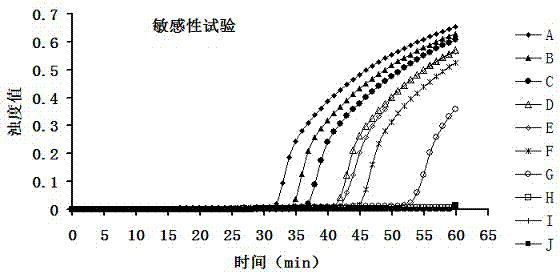
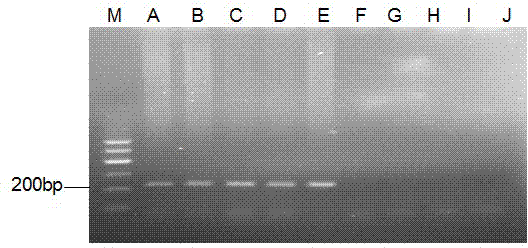
The invention discloses a streptococcus agalactiae loop-mediated isothermal amplification kit and application thereof. The kit comprises an LAMP primer, a 2*reaction buffer solution, Bst DNA polymerase, a fluorescent visual detection reagent, ultrapure water and a streptococcus agalactiae DNA template, wherein the LAMP primer comprises outer primerS F3 and B3 and inner primers FIP and BIP. Specificity detection and sensitivity detection prove that the LAMP detection kit disclosed by the invention can be used for monitoring reaction in real time, quantitatively detecting the copy number of the streptococcus agalactiae, rapidly and accurately obtaining detection results and providing convenience for simply and rapidly detecting the streptococcus agalactiae.
Owner:GUANGXI ACADEMY OF FISHERY SCI
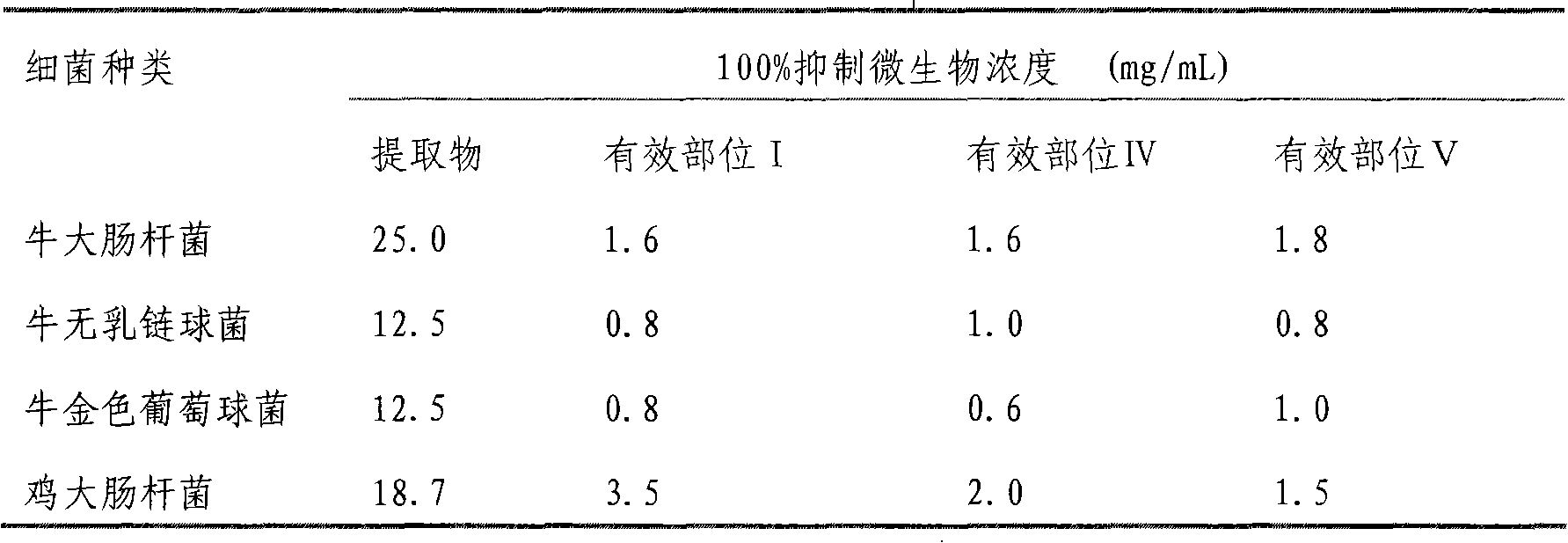
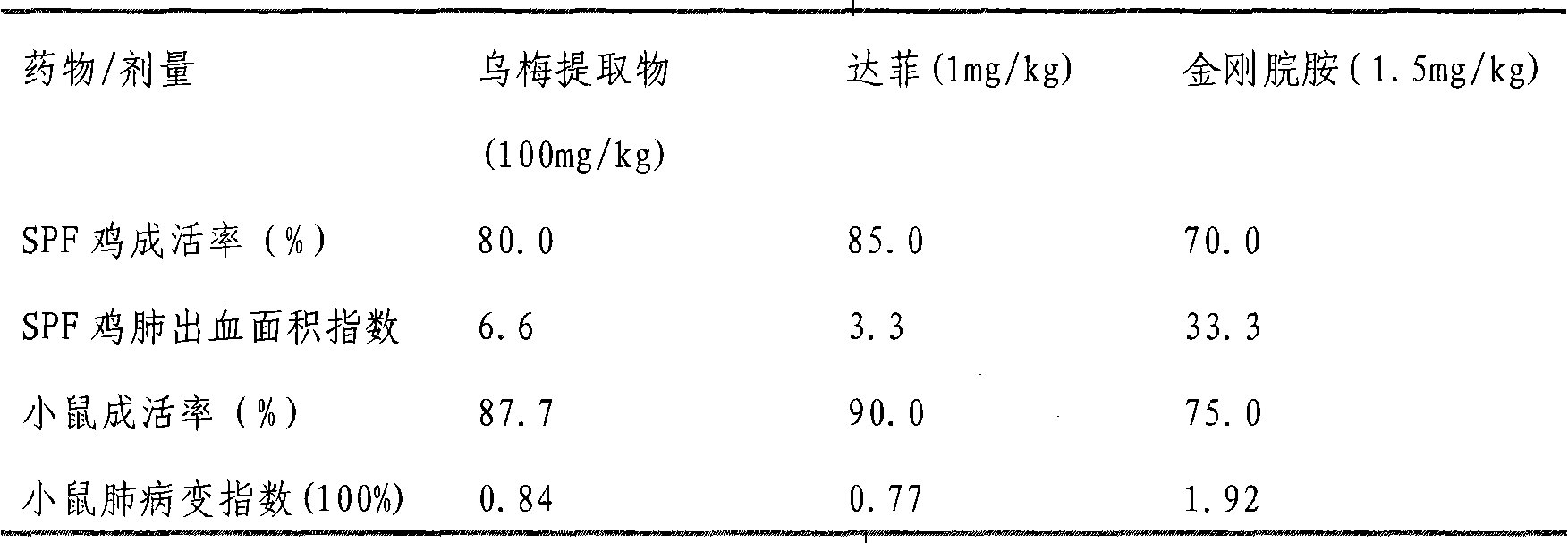
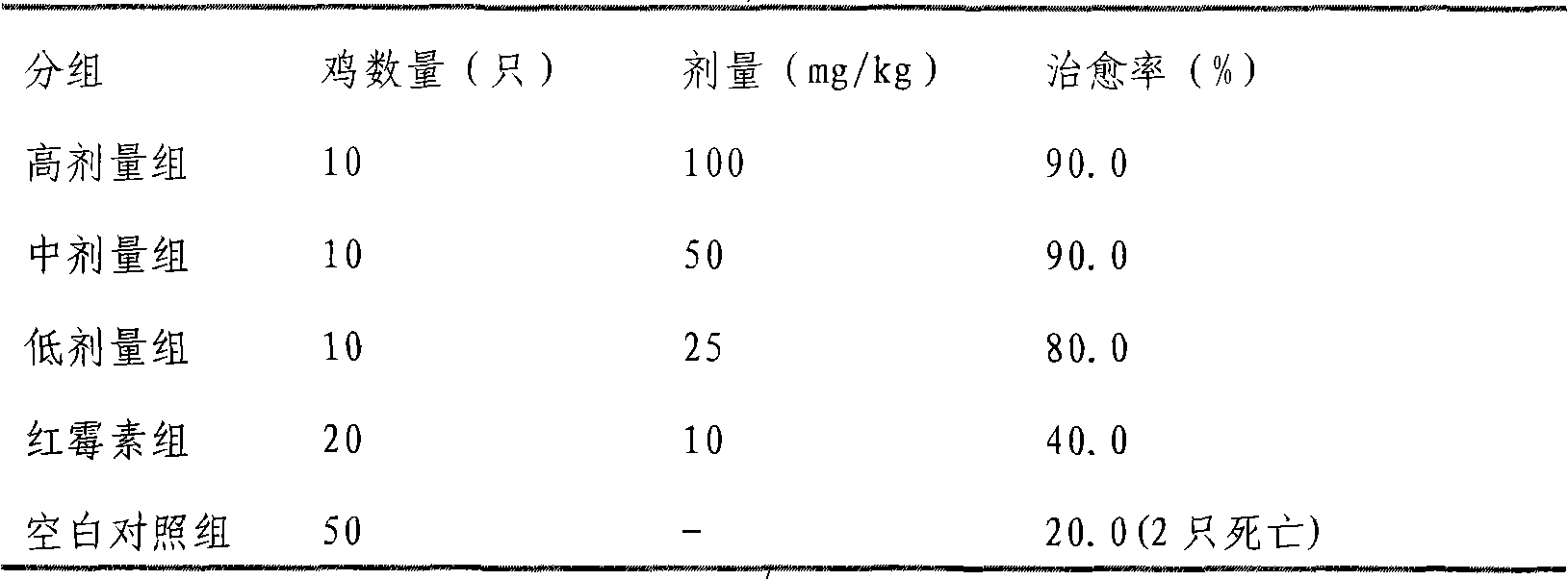
Uses of smoked plum extract against virus, bacteria, mycoplasma or chlamydia of livestock and poultry
ActiveCN101317904APrevent fluPrevent proliferationAntibacterial agentsAntiviralsBacteroidesSmoked Plum
The present invention discloses the application of ebony extract or effective parts (I, II and III) in preventing and remedying livestock and poultry viral disease, bacterial disease, mycoplasma disease and chlamydia disease. Tests in vitro and in vivo show that the ebony extract or effective parts (I, II and III) have the effects of preventing and inhibiting poultry flu virus, newcastle disease virus, infective bronchitis virus, and the like, and can effectively kill pathogenic bacteria causing the animal disease at the same time, such as bovine streptococcus agalactiae, bovine gold staphylococcus, bovine colon bacillus, chicken colon bacillus, etc. In addition, the ebony extract or effective parts can effectively prevent and remedy the poultry chlamydia disease and mycoplasma disease.
Owner:何诚
Preparation method and application of recombinant lactococcus lactis
ActiveCN107653260AImprove infection abilityAntibacterial agentsBacterial antigen ingredientsVector vaccineSerotype
The invention relates to a preparation method and application of a recombinant lactococcus lactis. The invention is characterized in that the surface immunogenic protein (Sip) of tilapia-source streptococcus agalactiae is intracellularly or secretorily expressed in lactococcus lactis cells, vectors which are used for the recombinant expression of the Sip are pNZ8124 and pNZ8148 respectively, signal peptide segments are removed from Sip gene segments inserted into the vectors, and histidine sequence tags are added to the Sip gene segments, the recombinant protein is induced to be expressed by Nisin, the optimum induction condition is 4h of induction by 100ng / mL of Nisin, the optimum oral immunization concentration is 2.24*10<10>CFU / mL, and the oral dosage is 100 mu L. The recombinant lactococcus lactis disclosed by the invention which is applied in lactobacillus living vector vaccines against tilapia streptococcus agalactiae has the advantages of wide serotype coverage, direct oral administration, high safety, easiness in operation, easy large-scale herd immunity, good immune effect and the like.
Owner:PEARL RIVER FISHERY RES INST CHINESE ACAD OF FISHERY SCI
Tilapia mossambica source streptococcus agalactiae low virulent strain and application thereof
ActiveCN104152368AStrong immune protectionReduced toxicityAntibacterial agentsBacterial antigen ingredientsBiotechnologyDisease
The invention relates to an aquaculture animal disease control technology and in particular relates to a tilapia mossambica source streptococcus agalactiae low virulent strain and application thereof. The tilapia mossambica source streptococcus agalactiae low virulent strain is YM001 and is preserved in CCTCC (China Center For Type Culture Collection) with the preservation number of CCTCCM2014045. The tilapia mossambica source streptococcus agalactiae low virulent strain is subjected to activation culture and fermentation culture, bacterial cells of a 106-109CFU / mL low virulent strain YM001 and a sterile phosphate buffered solution are mixed as a low virulent live vaccine used for preventing and controlling a tilapia mossambica streptococcus disease, the injection medication concentration for each tilapia mossambica is 105-108CFU, the oral medication concentration for each tilapia mossambica is 108-109CFU, and the soaking medication concentration is 107-108CFU / mL. The invention relates to strain identification, toxicity test, toxicity return safety and stability, immunogenicity and immune protection effect of a low virulent strain, and the tilapia mossambica streptococcus disease vaccine prepared by adopting the low virulent strain is good in immune protection effect and strong in safety and stability.
Owner:GUANGXI ACADEMY OF FISHERY SCI
Latex agglutination kit for detecting streptococcus agalactiae antibodies and application of kit
ActiveCN103529204AStrong specificityHigh sensitivityDisease diagnosisImmunoassaysSerum igeStreptococcus agalactiae
The invention discloses a latex agglutination kit for detecting streptococcus agalactiae antibodies of dairy cows as well as a preparation method and an application of the kit. The kit comprises latex agglutination particles, positive control serum, negative control serum, a sample diluent, a glass slide and a graduated pipette. The kit is used for detecting whether the dairy cows suffer from mastitis, is simple, convenient and fast in method, has strong specificity and high sensitivity and is suitable for being used for detection by grass-roots veterinarians; and instruments and equipments are not needed.
Owner:武汉市畜牧兽医科学研究所
Method for screening candidate bacterial strain from fish streptococcus agalactiae vaccine
ActiveCN102676683AHigh immune protection rateLarge range of protectionMicrobiological testing/measurementMicroorganism based processesGenotype AnalysisBacterial strain
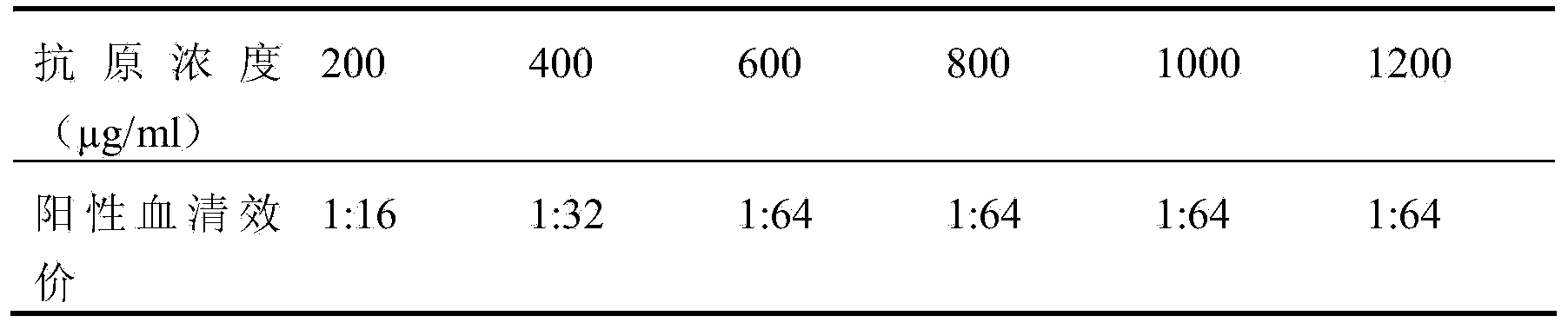
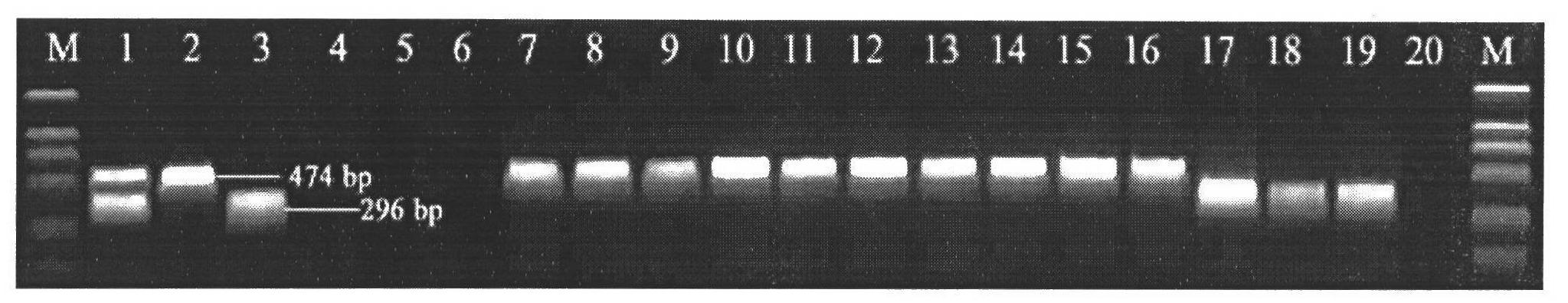
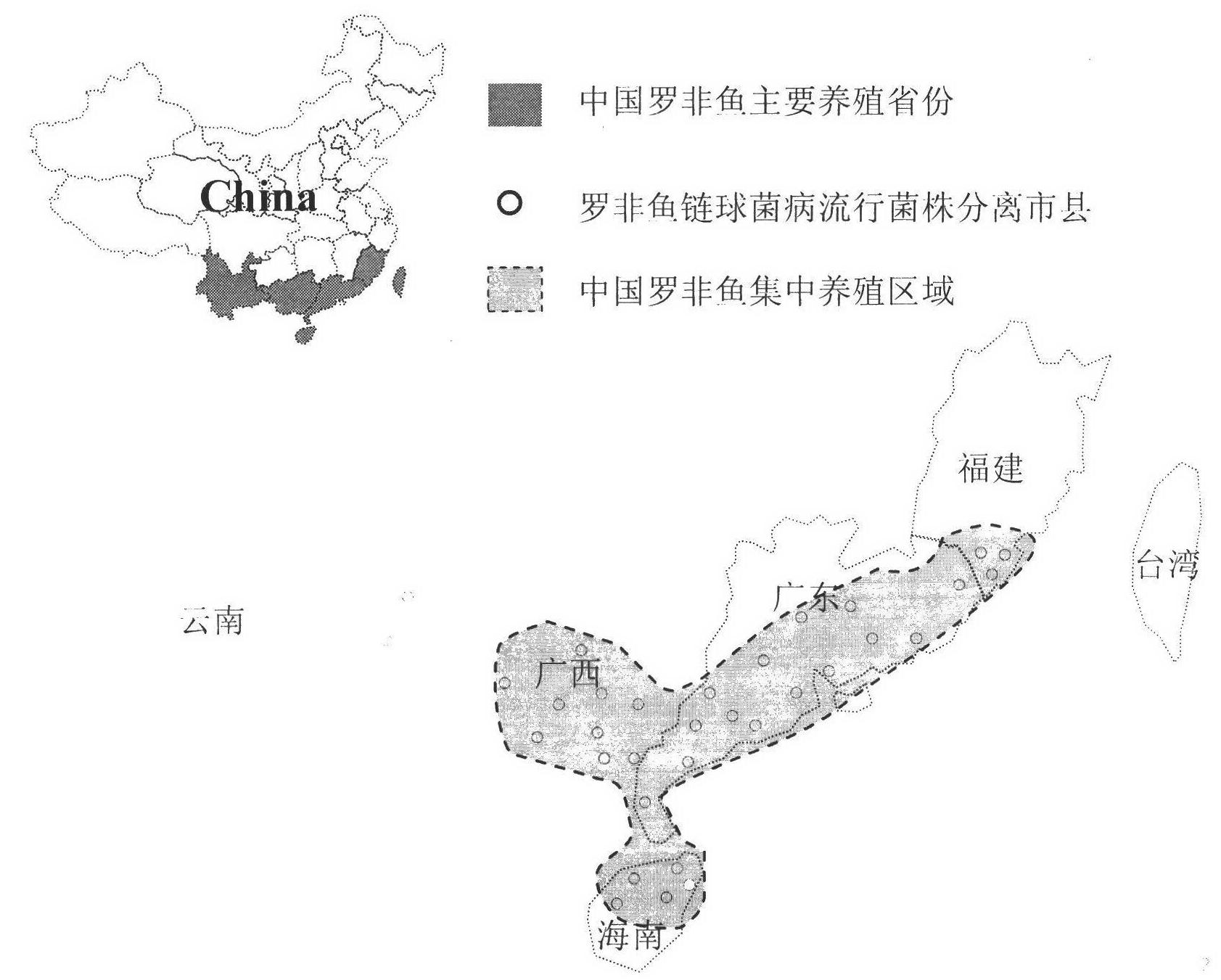
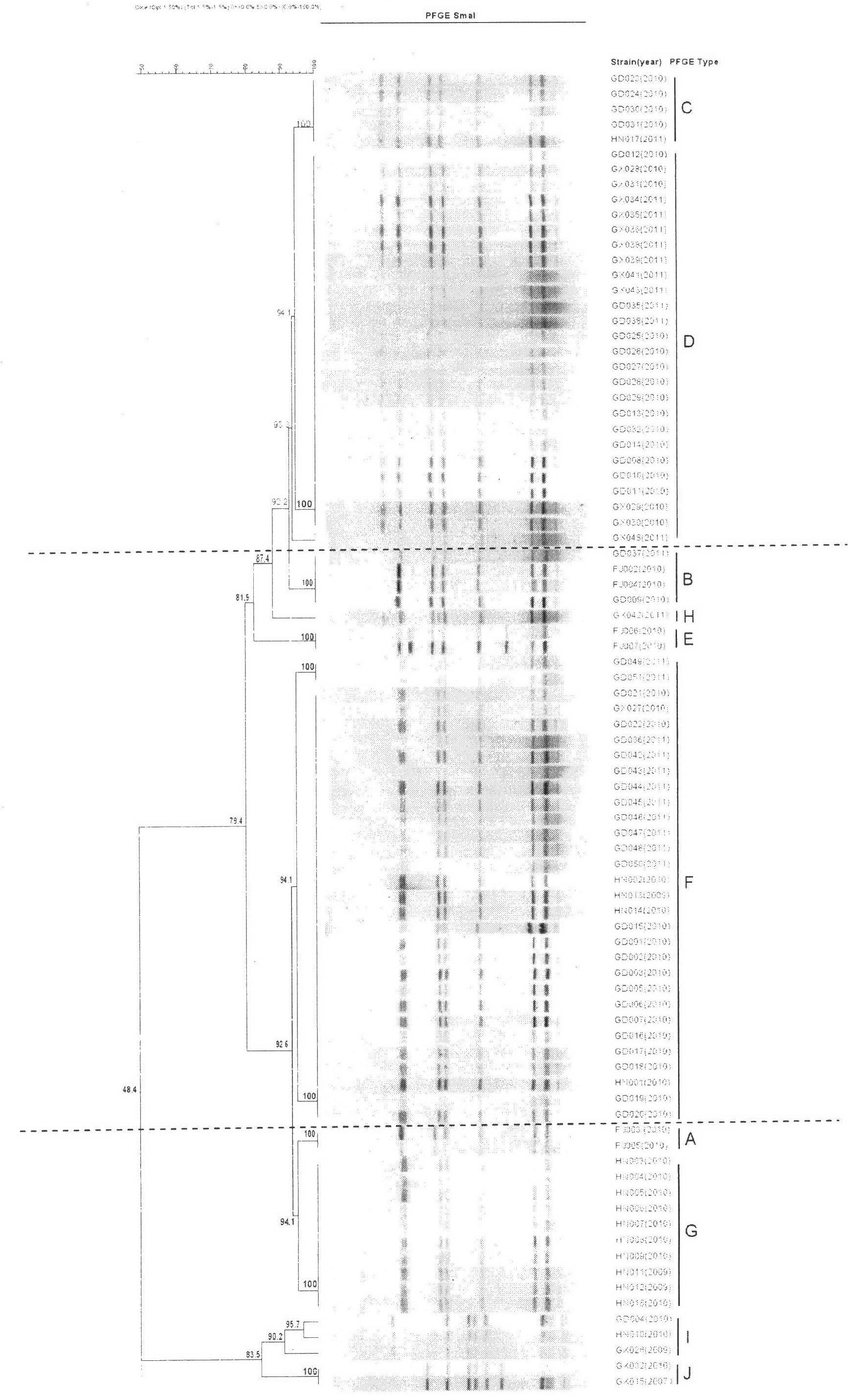
The invention discloses a method for screening candidate bacterial strains from fish streptococcus agalactiae vaccines. The method comprises the steps: separation and breed conservation of tilapia streptococcicosis agalactiae epidemic strains, identification of the epidemic strains and the establishing of a pathogenic library, PFGE genotype analysis of epidemic strains, toxicity determination of PFGE genotype representative strains, and immunogenicity and protection domain test of the PFGE genotype representative strains so as to obtain a candidate bacterial strains combination of a tilapia streptococcicosis agalactiae immunoprophylaxis vaccine in China. The technology has the characterizes of strong pertinence, high screening efficiency, remarkable effect and the like, and the screen candidate bacterial strains combination of the tilapia streptococcosis agalactiae immunoprophylaxis vaccine in China can protect 90% of genotypes and 96.47% of epidemic strains in the pathogenic library. The technology provides a new method for screening the candidate bacterial strains from the fish streptococcus agalactiae vaccines, is suitable for screening the candidate bacterial strains from the fish streptococcus agalactiae vaccines, and has significance and using value in immune prevention and control on fish streptococcicosis.
Owner:GUANGXI INST OF FISHERIES
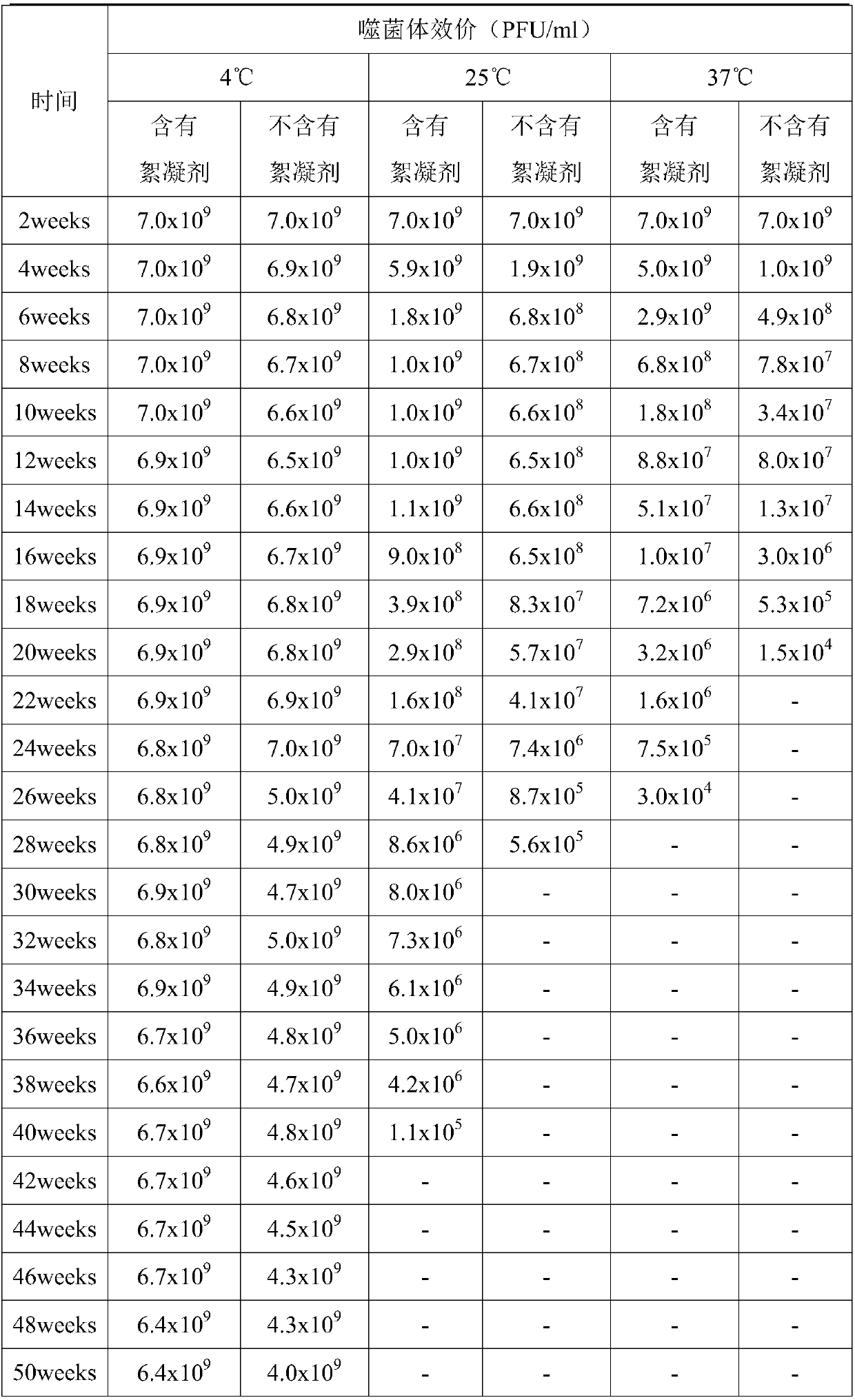
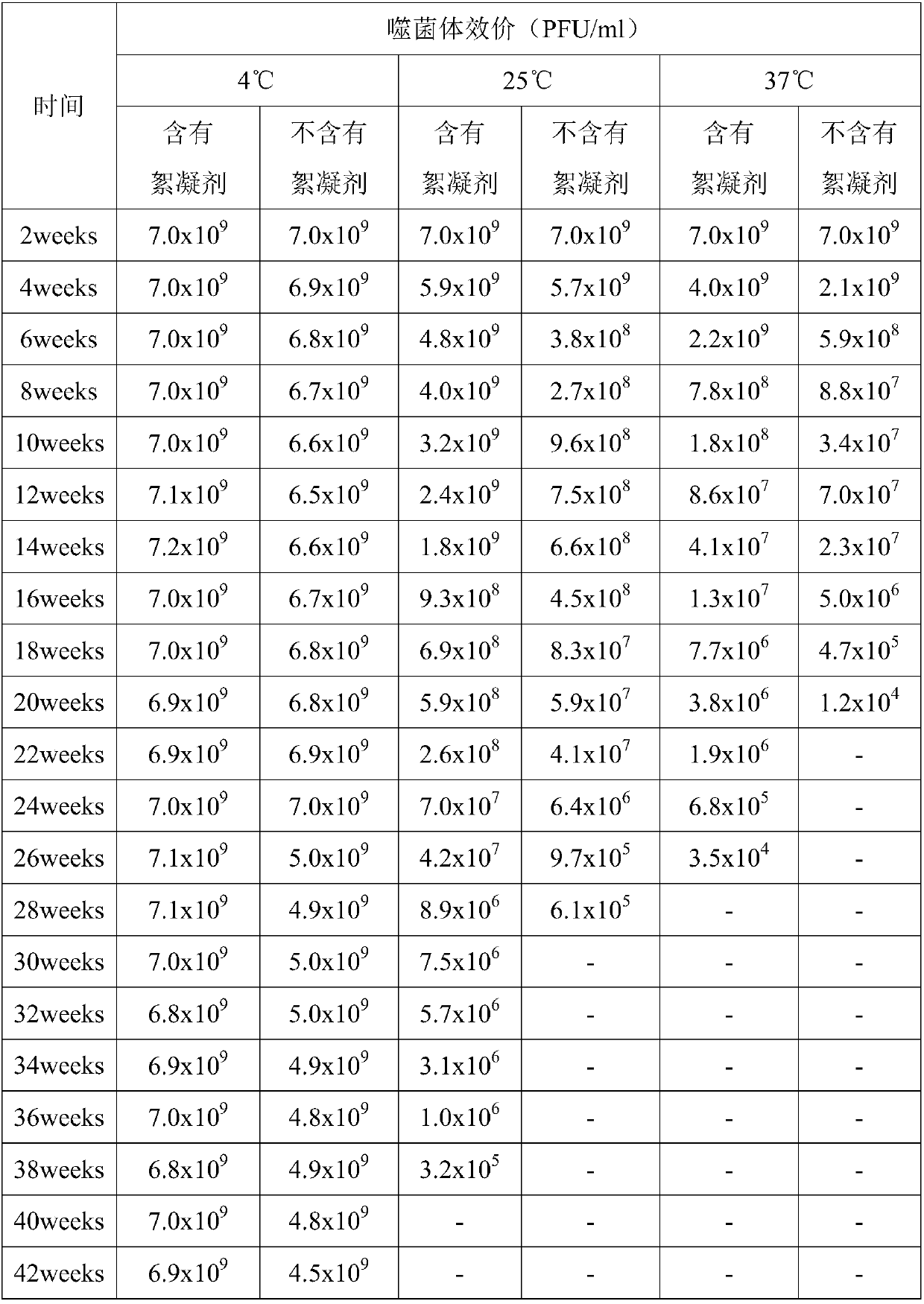
Phage flocculant and application of phage flocculant in fermentation post-treatment technology
PendingCN109593728ALow costAvoid elevationRecovery/purificationBacteriophagesBacteroidesXanthomonas axonopodis
The invention discloses a phage flocculant and application of the phage flocculant in a fermentation post-treatment technology and belongs to the field of a microbial post-treatment technology. The phage flocculant is characterized in that the phage flocculant is a cationic flocculant, and a working concentration of the phage flocculant is 0.1-5% generally. The flocculant is applied in the fermentation post-treatment technology of a vibrio parahaemolyticus phage, a coliphage, a salmonella phage, a xanthomonas axonopodis phage, a ralstonia solanacearum phage, a staphylococcus aureus phage, a streptococcus agalactiae phage, a streptococcus iniae phage or a pseudomonas aeruginosa phage. The phage flocculant can play a role of rapid subsidence of bacteria, be convenient and quick to operate, reduce centrifugal equipment investment and manpower and material resource loss and meet requirements of industrial mass production of the phage, and is safe, nontoxic and harmless to animals and plants.
Owner:PHAGELUX (NANJING) BIO-TECH CO LTD
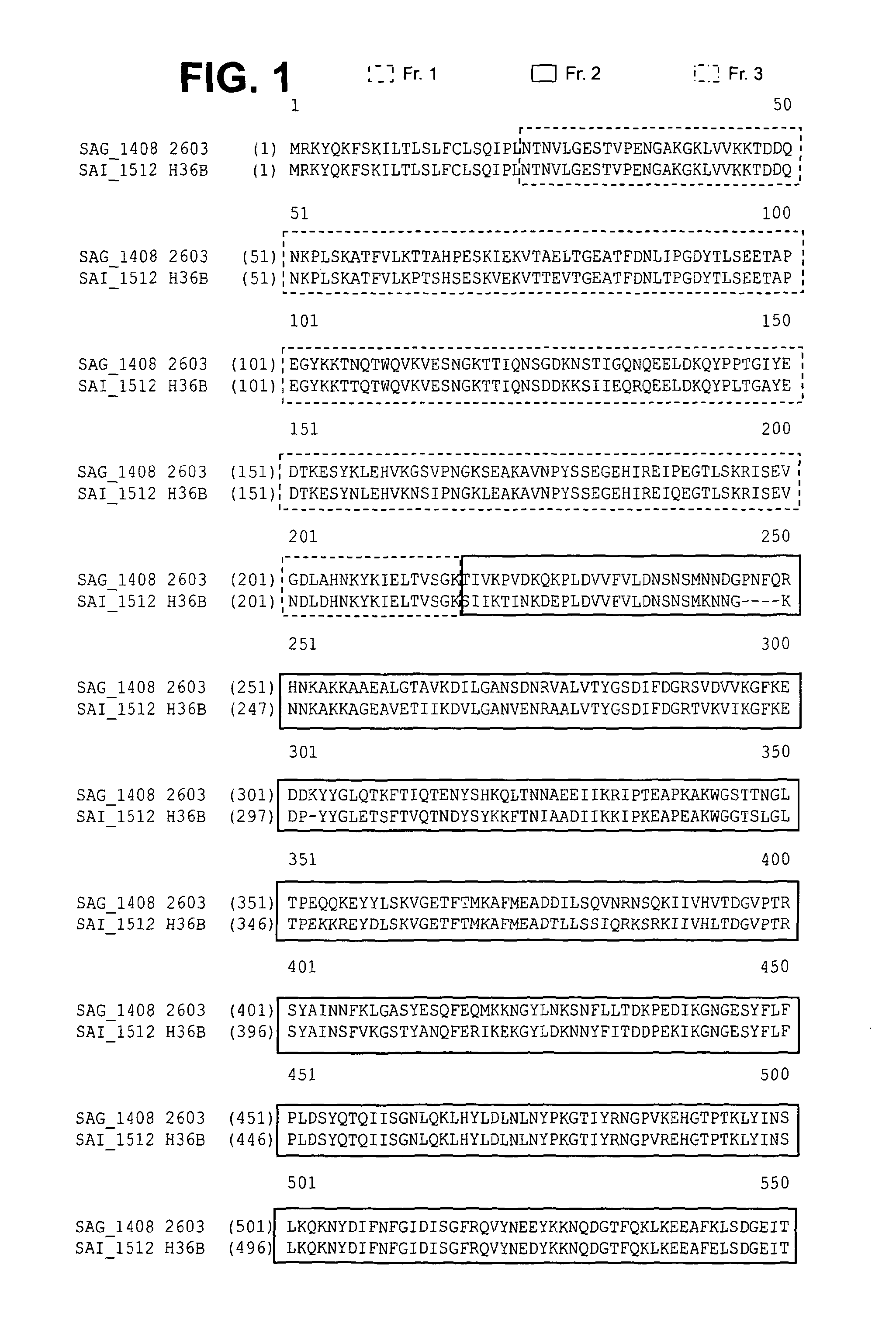
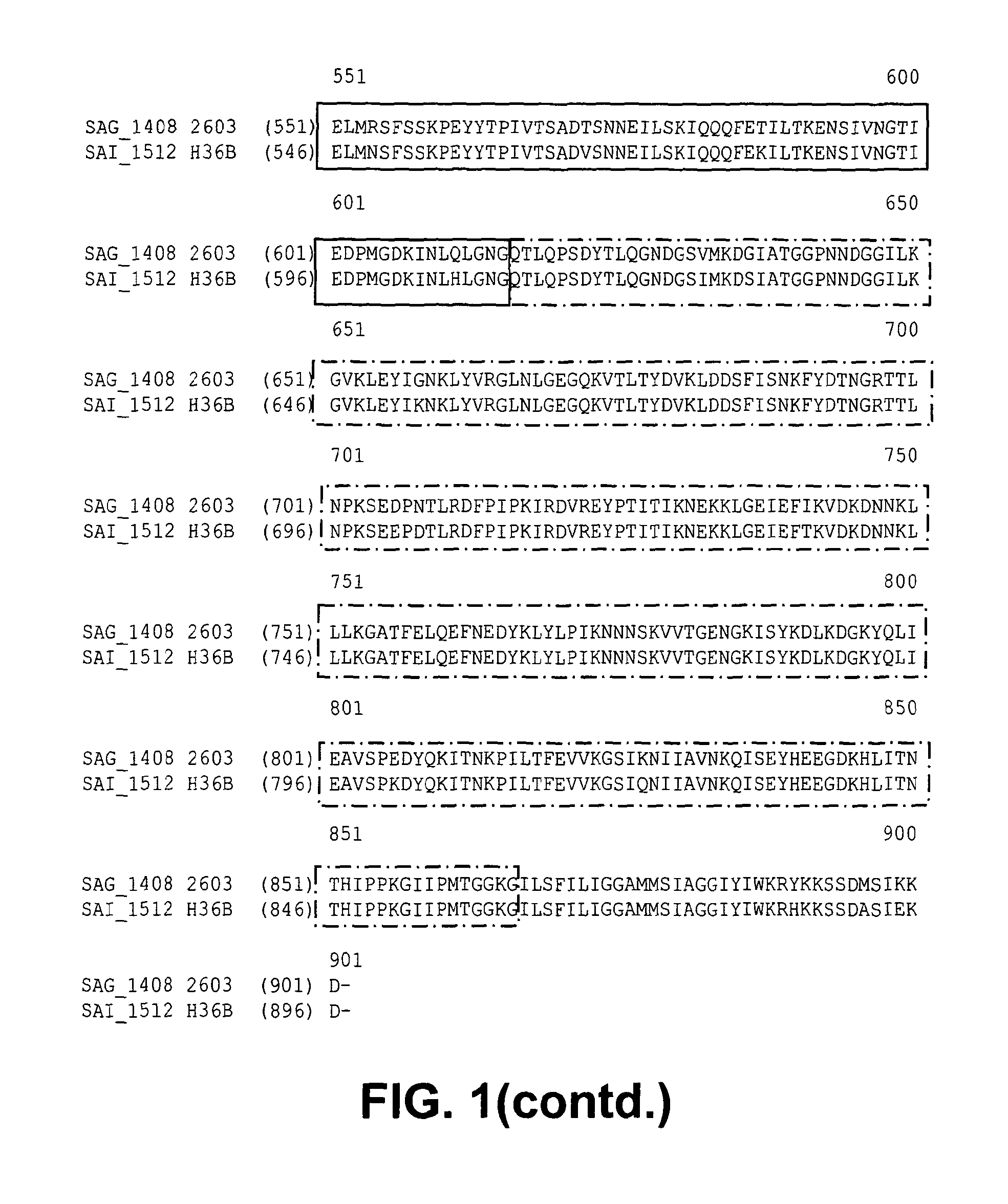
Immunogenic proteins and compositions
InactiveUS20130216568A1Enhance cellular immunityIncrease in antigen-specific CTL productionAntibacterial agentsPolypeptide with localisation/targeting motifStreptococcus agalactiaeMicrobiology
The invention provides proteins and compositions for the treatment and prevention of Streptococcus agalactiae (Group B streptococcus; GBS).
Owner:NOVARTIS AG
Group b streptococcus antigens
The present invention relates to polypeptides, epitopes and antibodies directed to these epitopes, more particularly to the Sip polypeptide of Group B streptococcus (GBS), also called Streptococcus Agalactiae which may be used to prevent, diagnose and / or treat streptococcal infection.
Owner:ID BIOMEDICAL
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com