Two step genome comparison method for designing quantitative PCR specific primer of guizhouense NJAU 4742
A comparative design and genome technology, applied in the field of agricultural microorganisms, can solve the problems of ungenome data update, information lag, time-consuming and labor-intensive, etc., and achieve the effect of good quantitative specificity and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Embodiment 1 is tested bacterial strain source
[0024]The Guizhou Trichoderma NJAU 4742 (preserved in the General Microbiology Center of the China Microbial Strain Preservation Management Committee, the preservation date is April 11, 2016, and the preservation number is CGMCC NO.12166) tested in the present invention and the Agrobacterium-mediated The gfp-labeled NJAU 4742 strain gfp-NJAU 4742 was provided by Jiangsu Provincial Key Laboratory of Solid Organic Waste Recycling High-Tech Research. The construction of the gfp strain is briefly described as follows: T -The DNA was inserted into the NJAU 4742 genome, and the successfully marked and stably inherited Trichoderma gfp-NJAU 4742 strain was obtained, and the inserted fragment was verified as a single copy by southern blotting experiments. Finally, the combination of Biolog analysis and plate confrontation test results of pathogenic fungi shows that the fluorescently labeled strain has the same phenotypic character...
Embodiment 2
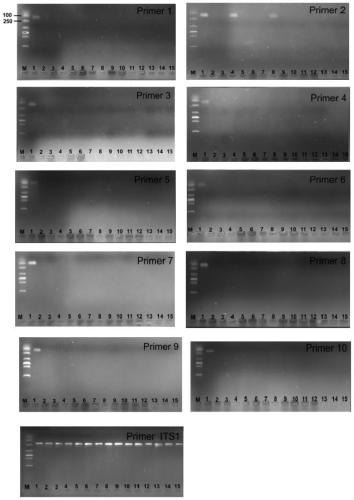
[0029] Example 2 Primer sequence selection and cross-amplification verification
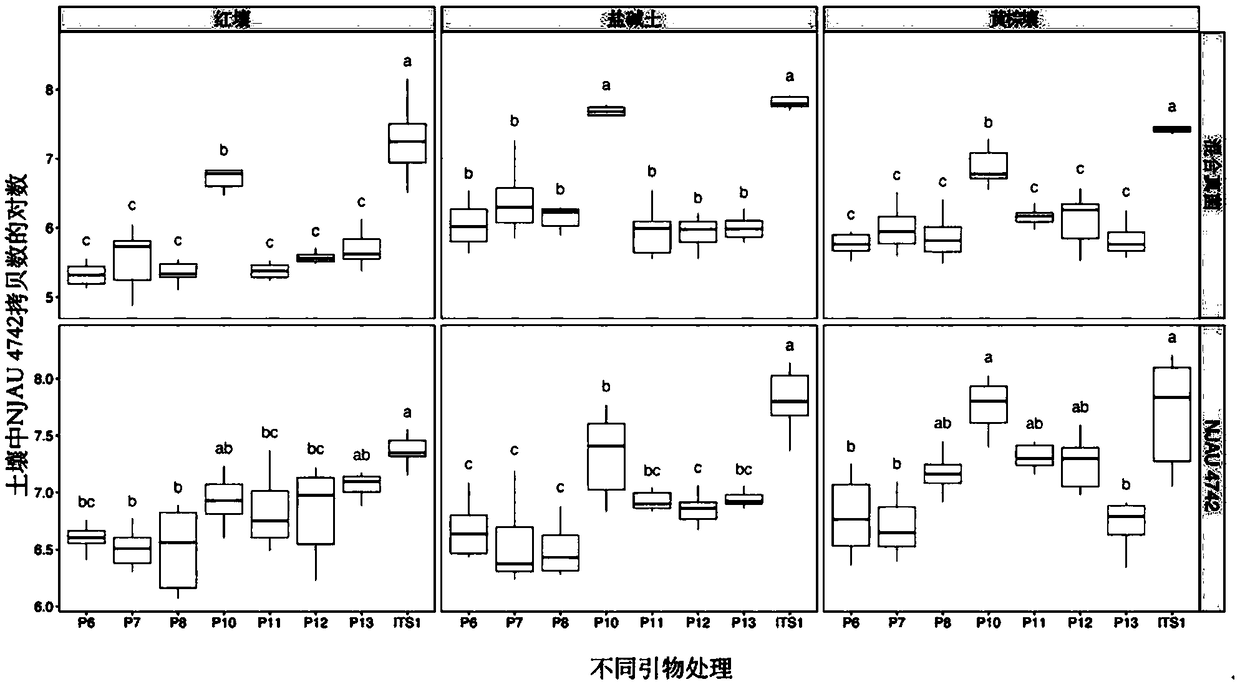
[0030] The present invention uses a two-step comparison method to compare the whole genome sequences of two strains with similar compatriots based on the database to find the specific sequence only for NJAU 4742. Step 1: Screen and compare the whole genome sequence of NJAU 4742 in the two authoritative databases of NCBI and JGI, and select genome fragments larger than 500bp that do not match. Step 2: compare the screened fragments with the whole genome of the compatriot strain 916 one by one, and obtain a series of unique base sequences that specifically distinguish the strain 916 as the source of the primer sequence design for the whole genome of NJAU 4742. At the same time, the present invention also selects the single-copy fragment inserted in the gfp-marker strain gfp-NJAU 4742 mediated by Agrobacterium to design three pairs of primers as the test standard for the specificity of the primers d...
Embodiment 3
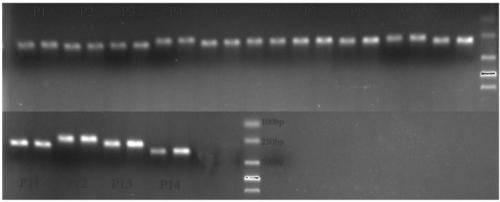
[0035] Construction and quantitative PCR of embodiment 3 plasmid
[0036] 10 pairs of primers designed from the selected target gene region in the genome of Trichoderma Guizhou NJAU 4742, and 3 pairs of primers designed for the gfp insert (two pairs of primers P11 and P13 are designed to be related to the gfp gene, and P12 is designed to be connected to the GFP protein. A section of hygromycin gene, which is used for screening marker strains, the hygromycin fragment and GFP are connected as a whole) and the fragment amplified by primer ITS1 (Table 2) are respectively cloned into the pMD19-T vector (TaKaRa) , and transform the plasmid into E. coli Top10 cells, use the PMD19-T universal sequence primer M13-F: TGTAAAACGACGGCCAGT (SEQ ID NO.43) and M13-R: CAGGAAACAGCTATGACC (SEQ ID NO.44) for PCR amplification The fragments in the plasmid were verified and sequenced by Nanjing GenScript Biotechnology Co., Ltd. The DNA concentration of the plasmid was measured using a spectrophoto...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com