Specific primer for novel coronary virus detection and rapid detection method
A coronavirus, detection method technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, resistance to vector-borne diseases, etc. and other problems, to achieve the effect of good sequencing quality, rapid and accurate detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
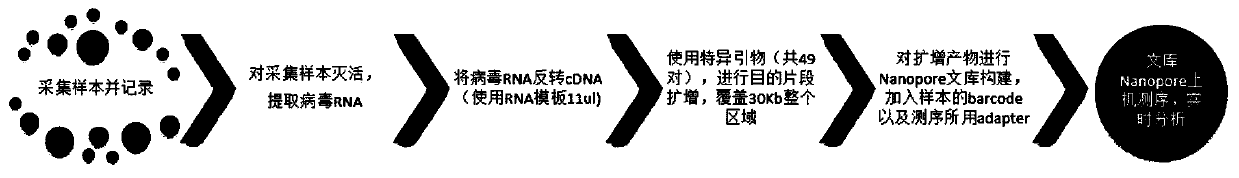
[0045] Such as figure 1 as shown, figure 1 It is a schematic flow diagram of the rapid detection method of the novel coronavirus; the rapid detection method of the novel coronavirus of the present invention comprises steps:
[0046] S1, inactivate the collected samples and extract viral RNA;
[0047] Virus inactivation treatment method: According to literature and guideline reports, coronavirus can be inactivated at 56°C for 30 minutes. Our preliminary tests show that the inactivation method of dry bath (oven or incubator) at 56°C for 30 minutes or 50°C for 60 minutes does not affect respiratory samples. Detection of RNA viruses. Preferably, respiratory samples (except for the purpose of cultivating microorganisms) are first dry bathed at 56°C for 60 minutes, allowed to stand for 10 minutes to room temperature (to avoid aerosols), and then processed.
[0048]Viral RNA extraction and quality inspection: The collected biological samples, including throat swabs, alveolar washi...
Embodiment 2
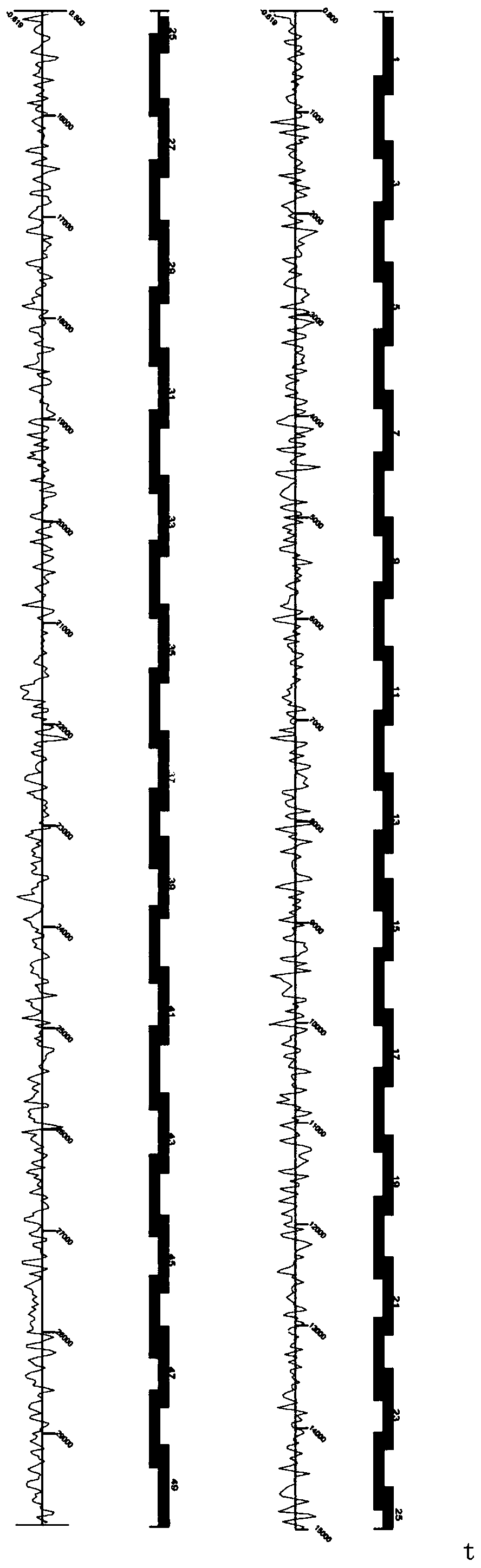
[0092] In this embodiment, for a highly suspected patient, but the three PCR results are all negative, such as Figure 4 as shown, Figure 4 It is the result graph of the three PCRs of the patient. However, when the rapid detection method of the present invention is used for detection, a large number of novel coronavirus genome sequences are observed in the patient's extracted samples, such as Figure 5 as shown, Figure 5 A map of the genome sequence alignment of the sample extracted for the patient. Figure 5 The first row is the reference genome of the new coronavirus, and a large number of high-coverage and uniform alignment sequences appear at the bottom. This result is positive through serological IgM test samples, which can verify that the patient is carrying the new coronavirus. As shown in Table 2, Table 2 is a list of the virus detected in this patient.
[0093] Table 2 List of detected viruses
[0094]
[0095] Such as Image 6 , Figure 7 as shown, Imag...
Embodiment 3
[0107] The present invention can be applied to the sample DNA comparison results at the same time, as shown in Table 5, Table 5 is the detection list of the Neisseriaelongata, Actinomycesgraevenitzii and Prevotellamelaninogenica sequences found in the sample in Example 2, which may be related to the actual operation of the sample Sometimes there is a small amount of pollution.
[0108] Table five
[0109]
[0110] The detected results of the present invention can be compared with homologous sequences of other species. Such as Figure 14 as shown, Figure 14 It is the result of simultaneous comparison between the sample of Example 2 and the new crown and SARS. The READs compared to the new crown are more than the READs compared to the SARS. At the same time, most of the READs compared to the SARS are also compared to the new crown. This shows that SARS There are many similarities with the pathogenic region of the new crown.
[0111] Among all the samples in Example 2, th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com