Specific gene sequence of colletotrichum fructicola and application of specific gene sequence
A gene sequence, anthracnose technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve the problems of high professional background requirements for instruments, equipment and analysts, time-consuming and laborious, cumbersome steps, etc., and achieves simple and rapid identification steps. Reproducible, low-cost results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
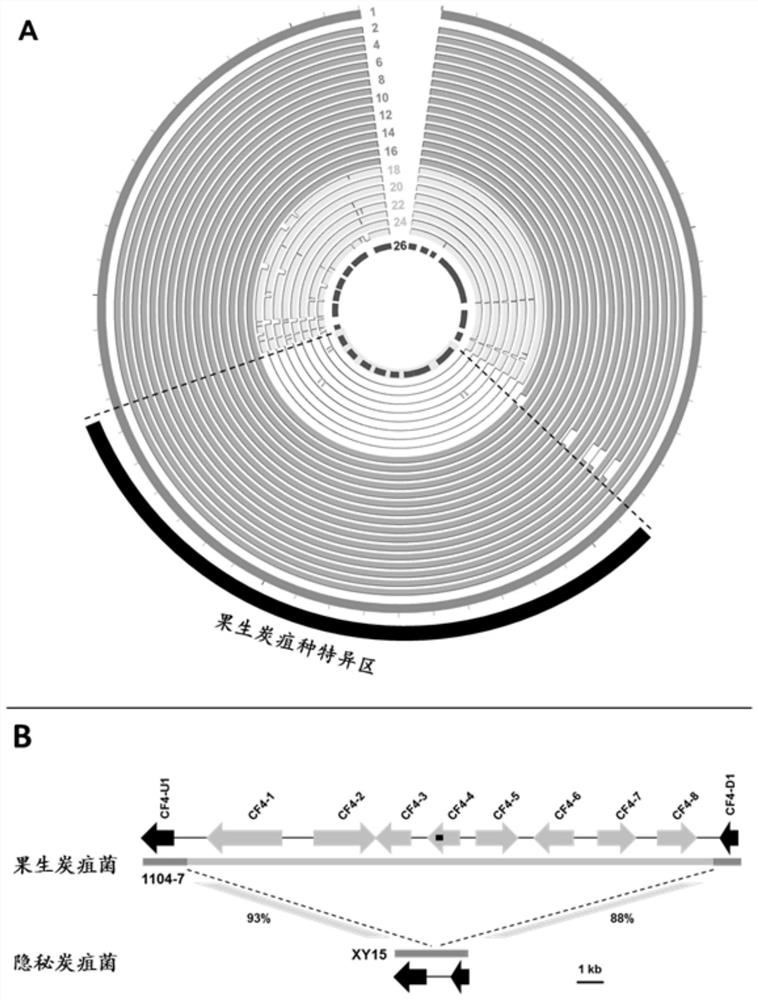
[0027] The species-specific DNA sequence of the anthracnose fruit of the present invention is 19878bp long, and its sequence is shown in SEQ ID No.1, and its acquisition and identification method is described as follows:
[0028] (1) Select more than 20 Ascomycete genomes in the GenBank public database, and perform OrthoMCL clustering on the encoded proteins of the entire genome, and screen out OrthoMCL clusters specific to the fruiting anthracnose species;
[0029] (2) Relying on the G. anthrax omics resources (17 genomes of fruiting anthracnose strains and 8 genomes of fruiting anthracnose relatives) obtained by the previous sequencing of the fungal research laboratory of Northwest Agriculture and Forestry University, OrthoMCL specific for the obtained fruiting anthrax Cluster clusters were analyzed by tBlastn and genome comparison analysis to verify the species specificity of the gene and the conservation among strains within the species of Anthrax fruit;
[0030] (3) On th...
Embodiment 2
[0041] This embodiment is an example of the development of the rapid detection kit for fruit-born anthracnose species. The kit of this embodiment includes conventional PCR reagents (commercialized kit AP111 of Beijing Quanshijin Biotechnology Co., Ltd.), and the sequence is SEQ ID No.2 Primer P1-F and primer P1-R whose sequence is SEQ ID No.3.
[0042] The specific steps of using the kit for rapid detection of fruit anthracnose are as follows:
[0043] (1) extracting the genomic DNA of the strain to be tested;
[0044] (2) DNA amplification: PCR amplification system: 10xEasy Taq Buffer 2.5μL, 2.5mM dNTPs 2μL, 10μM forward primer (SEQ ID NO.2) 0.5μL, 10μM reverse primer (SEQ ID NO.2) 0.5 μL, 1.0 μL of 100ng / μL DNA template, 0.2 μL of Easy Taq DNA polymerase, and make up to 25 μL with ddH2O. The PCR reaction program is: pre-denaturation at 94°C for 2 minutes; denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min, 40 cycles; extension at 72°C fo...
Embodiment 3
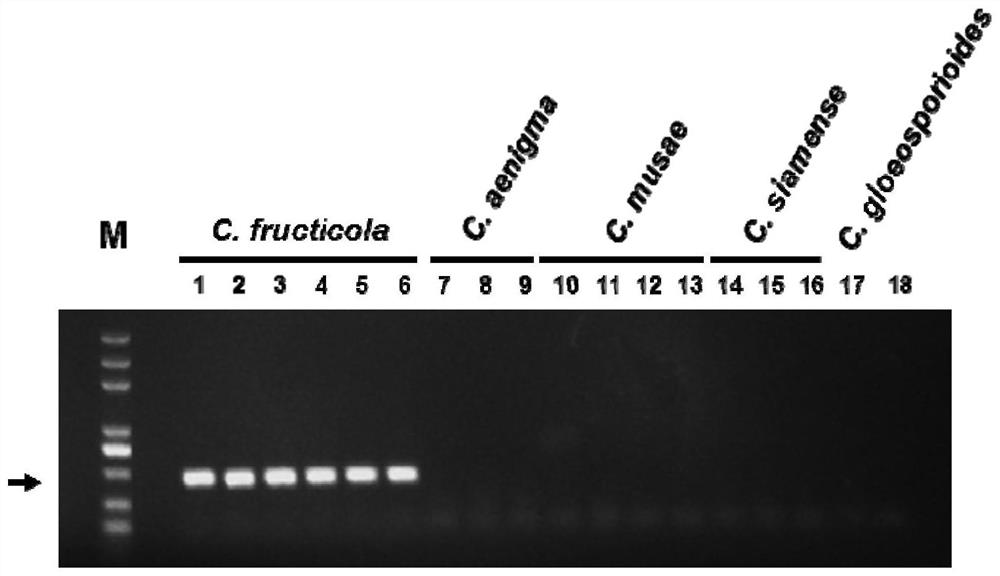
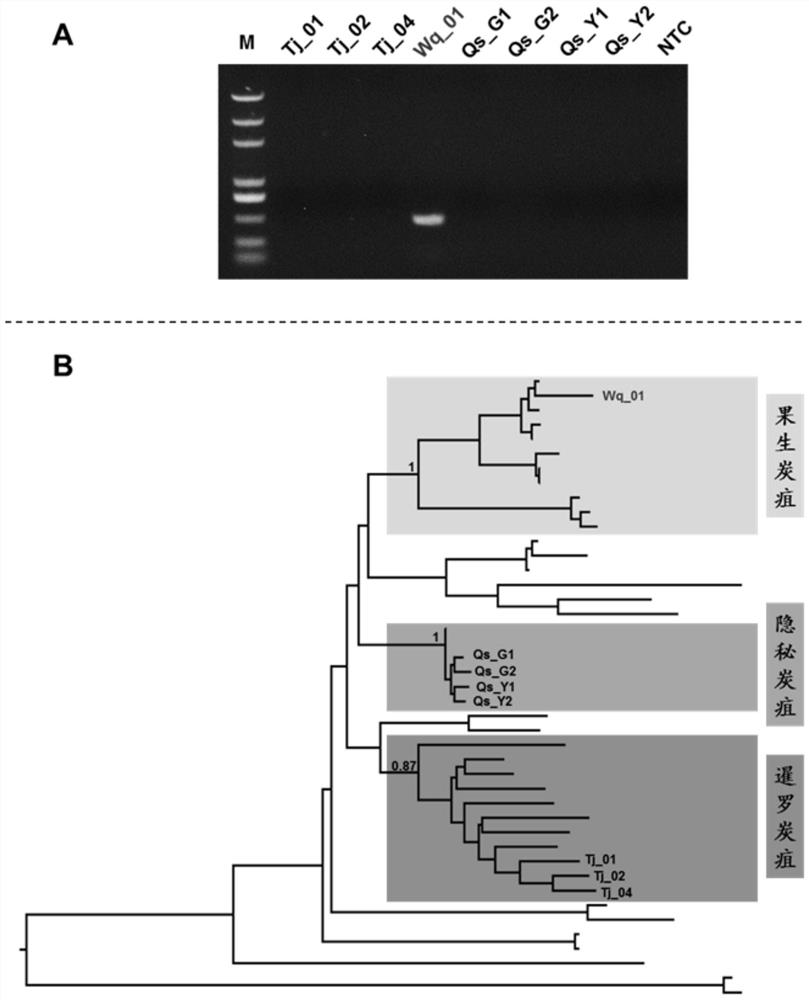
[0047] This embodiment shows that the rapid detection kit of fruit anthracnose bacteria (C. fructicola), secret anthracnose bacteria (C.aenigma), banana anthracnose bacteria (C.musae), Siamese anthracnose bacteria (C.siamense) ), the PCR amplification detection results of species such as C. gloeosporioides. details as follows:
[0048] Test strains: 17 test strains were used in this embodiment, including 6 fruit anthracnose bacteria, 3 cryptic anthracnose bacteria, 4 banana anthrax bacteria, 3 Siamese anthrax bacteria, and 1 glyospora anthracnose bacteria. All strains are stored in the China Agricultural Microorganism Culture Collection Management Center or the Fungal Laboratory of Northwest A&F University. The species classification information of the strains is confirmed based on the polygenetic phylogenetic analysis. The specific information of the strains is shown in Table 1:
[0049] Table 1 Example 3 test strain information table
[0050] serial number stra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com