Automated analysis method for prokaryotic transcriptome based on second generation sequencing
An automated analysis and next-generation sequencing technology, applied in the field of high-throughput sequencing, can solve problems such as unsatisfactory standardized results, waste of labor and time, and inaccurate differential analysis results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0030] The present invention will be further described by the following examples, but these examples should not be used to explain the limitation of the present invention.
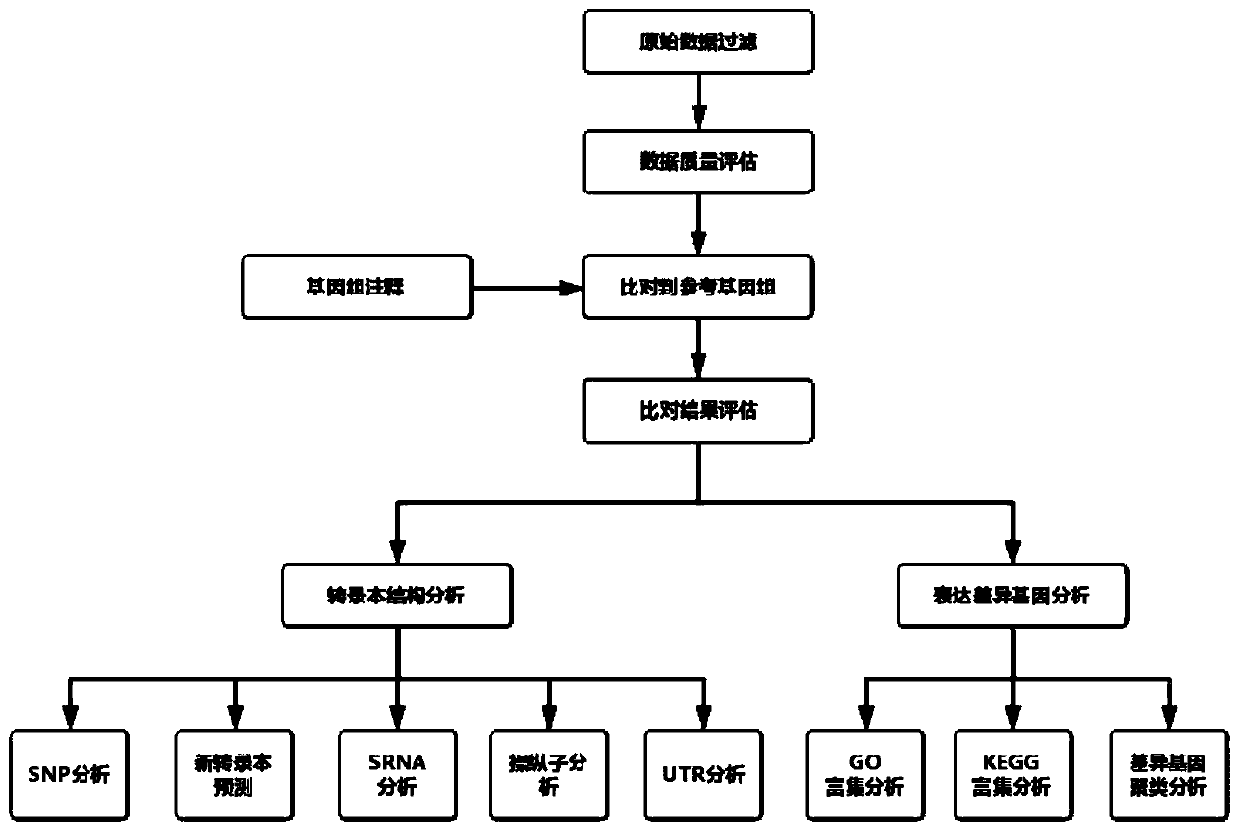
[0031] see process figure 1 , the steps of the present invention include:
[0032] S1) Prepare and read the config file, which includes: off-machine data location and corresponding sample name and group name, grouping for differential analysis, path for saving analysis results, task name, genome sequence and its index file location, GTF files and other information. When the software reads the relevant information, it will generate shell scripts corresponding to all the analysis steps listed below, and run them in order. There will be a running log for each step while running, which is convenient for result checking. All analytical steps are briefly described below.
[0033] The first is raw off-board data filtering. As an example, use the perl script independently developed by Pysennuo to filter the or...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com