Streptococcus agalactiae attenuated strain YM001 genome sequence feature and use thereof
A genome sequence and Streptococcus galactiae technology, applied in the field of Streptococcus agalactiae genome sequence, can solve the problems of indistinguishable, difficult to maintain intellectual property rights and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0010] Example 1 Whole Genome Sequencing and Sequence Characteristics of Streptococcus agalactiae Attenuated Strain YM001
[0011] Genomic DNA of the attenuated Streptococcus agalactiae strain YM001 was extracted, and the library was constructed after passing the test. First, the genomic DNA was randomly interrupted by the ultrasonic method Covaris or Bioruptor to generate a series of DNA fragments with a main band less than or equal to 800bp, and then the sticky ends formed by the interruption were repaired into blunt ends by T4DNAPolymerase, KlenowDNAPolymerase and T4PNK. The base "A" allows the DNA fragment to be connected to a special adapter with a "T" base at the 3' end, and the target fragment ligation product to be recovered is selected by electrophoresis, and then PCR technology is used to amplify the adapter at both ends. DNA fragments; finally, cluster preparation and sequencing with qualified libraries. Basic biological information analysis after disembarkation (1...
Embodiment 2
[0018] Example 2 Characteristics of CRISPRs of Gene Functional Elements of Streptococcus agalactiae Attenuated Strain YM001
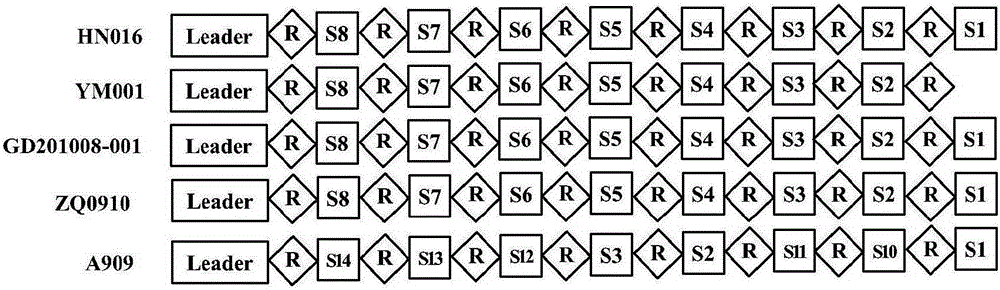
[0019] Searching and comparing the CRISPRs of 8 strains of Streptococcus agalactiae (see Table 2) (http: / / crispr.u-psud.fr / Server / ), the results showed that 3 strains of Streptococcus agalactiae serotype Ib were derived from tilapia CRISPRs were not detected, and CRISPR1 was detected in all 5 strains of Ia serotype Streptococcus agalactiae, but CRISPR2 was not detected. The CRISPRs sequences of strains HN016, GD201008-001 and ZQ0910 were consistent and had 8 spacers, while the attenuated strain YM001 had only 7 spacers, and other sequences were consistent with wild strains (see figure 2 ). It shows that it is difficult to eliminate the spacer sequence of CRISPRs of Streptococcus agalactiae under the condition of normal subculture in the wild, but in the absence of the threat of exogenous nucleic acid invasion, the spacer sequence of CRISPRs will be lost...
Embodiment 3
[0022] Example 3 Characteristics of Prophages of Gene Functional Elements of Streptococcus agalactiae Attenuated Strain YM001
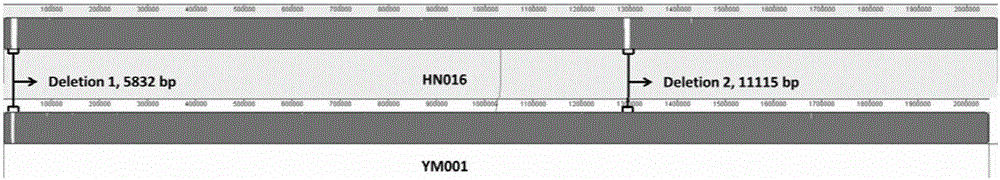
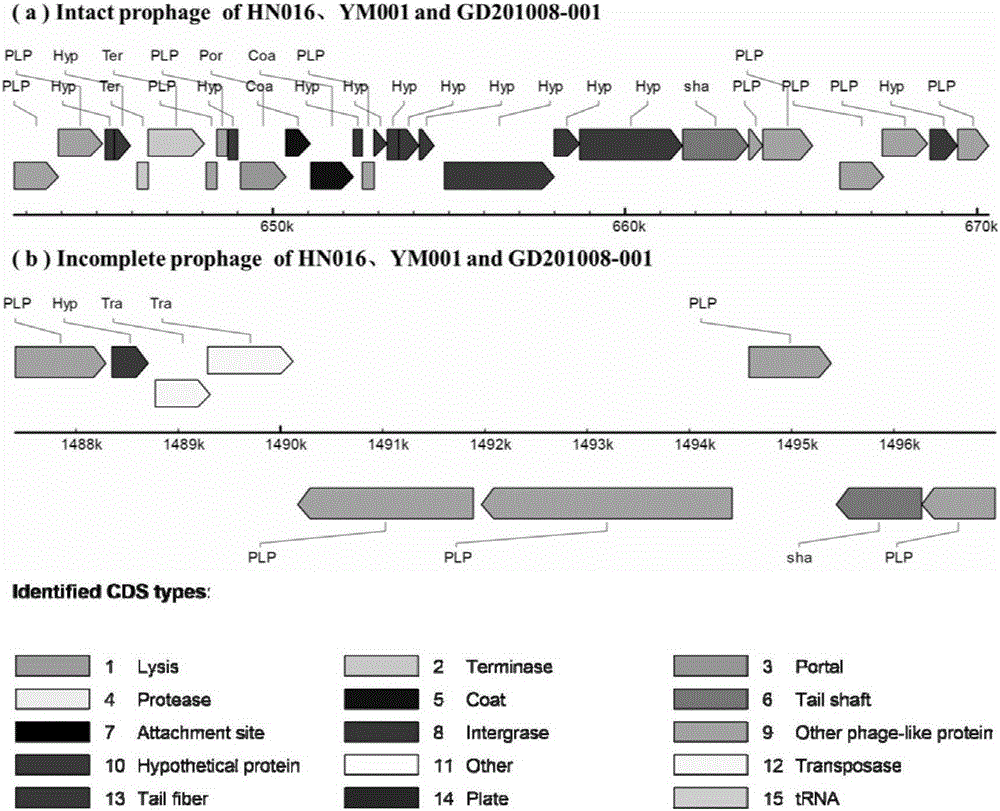
[0023] PHAST (http: / / phast.wishartlab.com / index.html) was used to analyze the prophages (Prophages) of HN016, YM001 and GD201008-001, and the results showed that a complete prophage sequence and 1 incomplete prophage sequence, and the coding sequence and position of all prophage sequences are completely identical (see image 3 ), indicating that the prophage sequence of the attenuated Streptococcus agalactiae strain YM001 has not changed, and the complete retention of the prophage sequence is conducive to the integrity of the attenuated strain's antigenicity.
[0024]
[0025]
[0026]
[0027]
[0028]
[0029]
[0030]
[0031]
[0032]
[0033]
[0034]
[0035]
[0036]
[0037]
[0038]
[0039]
[0040]
[0041]
[0042]
[0043]
[0044]
[0045]
[0046]
[0047]
[0048] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com