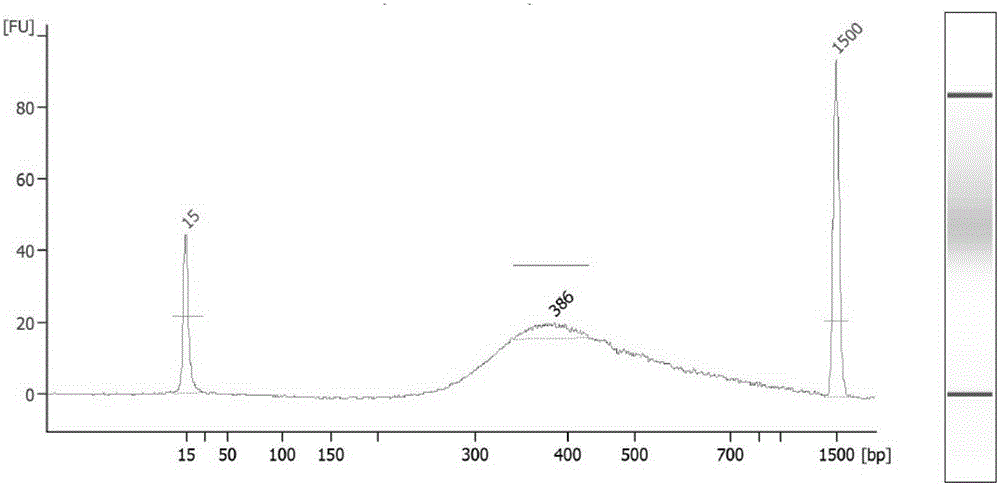
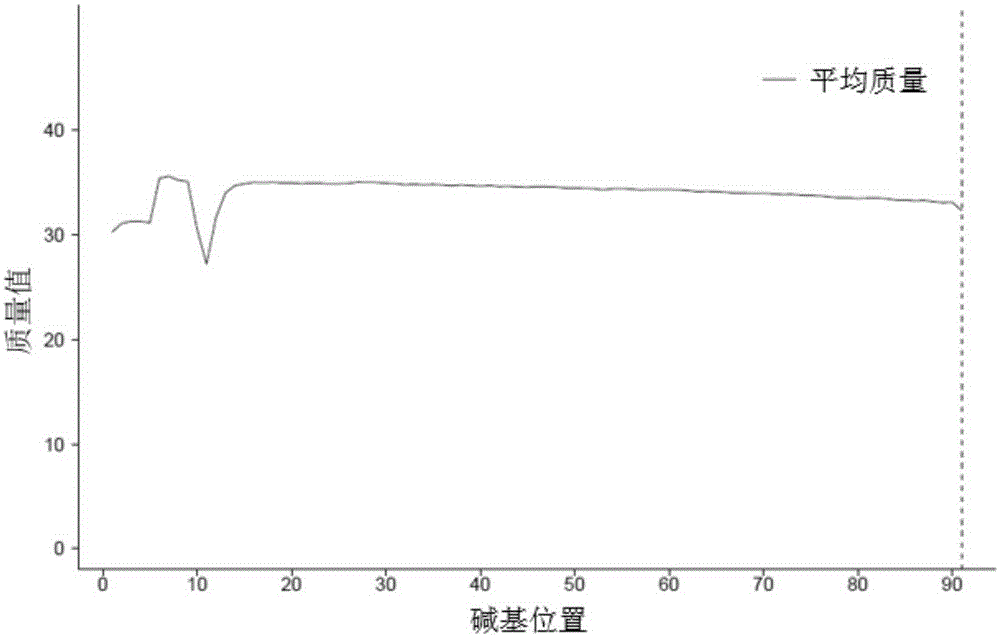
Method for acquiring chicken whole genome high-density SNP marker sites
A technology of whole genome and marker loci, which is applied in the field of obtaining high-density SNP marker loci in the whole genome of chickens, can solve the problems of interfering experimental results, affecting typing efficiency and cost, data loss, etc., and achieves technical stability and high repeatability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1 is used to illustrate the method of the present invention
[0042] 1. Experimental materials:
[0043] Collection of red native chickens, commercial layer breeds Bailaihang chickens, commercial broiler breeds Aiba Yijia broiler chickens, Lingnan yellow chickens, Chinese local chicken breeds Huiyang bearded chickens, Wenchang chickens, Henan game cocks, Qingyuan Ma chickens, black wolf pheasant chickens, Camellia Blood samples from 4-6 individual chickens, Peking chicken, Tibetan chicken, silky chicken, Shouguang chicken, bamboo silk chicken, Shiqi mixed chicken, Xianju chicken, invisible white chicken, and bantam yellow chicken, totaling 96 Individuals, the genome was extracted, and the genome concentration was diluted to 50ng / μL for use.
[0044] 2. Adapter and primer sequence:
[0045] Synthesize a pair of universal adapter sequences, 96 pairs of barcode adapter sequences, and a pair of PCR primer sequences.
[0046] 3. Sequencing library construction:...
Embodiment 2
[0068] The selection of embodiment 2 chicken genome optimum endonuclease combinations
[0069] Example 2 is used to illustrate the enzyme cleavage combination used in the present invention.
[0070] Considering the recognition characteristics of different restriction enzyme sites (such as the number of recognized bases, GC content, methylation status), etc., the inventor designed a total of 8 sets of double enzyme digestion combinations, and carried out different enzyme digestion through 3 individuals of Lingnan yellow chicken and Huiyang bearded chicken. The combined sequencing experiment, the experimental process is the same as that in Example 1, and the experimental results are shown in Table 2. It can be seen that the number of SNPs in the EcoRI–MseI digestion combination is 134,291 (the number of SNPs will vary with the number of experimental individuals), and the number of enzyme-digested fragments is 414,294, with the highest comparison rate with the genome. In one Next...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com