Method for quickly acquiring comparison result data of target genome region
A target gene and genome technology, which is applied in the field of quickly obtaining the data of target genome region comparison results, can solve the problems of reducing BAM storage, insufficient storage resources, lack of universality, etc., and achieves convenient operation and low computing resource requirements. , a wide range of effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
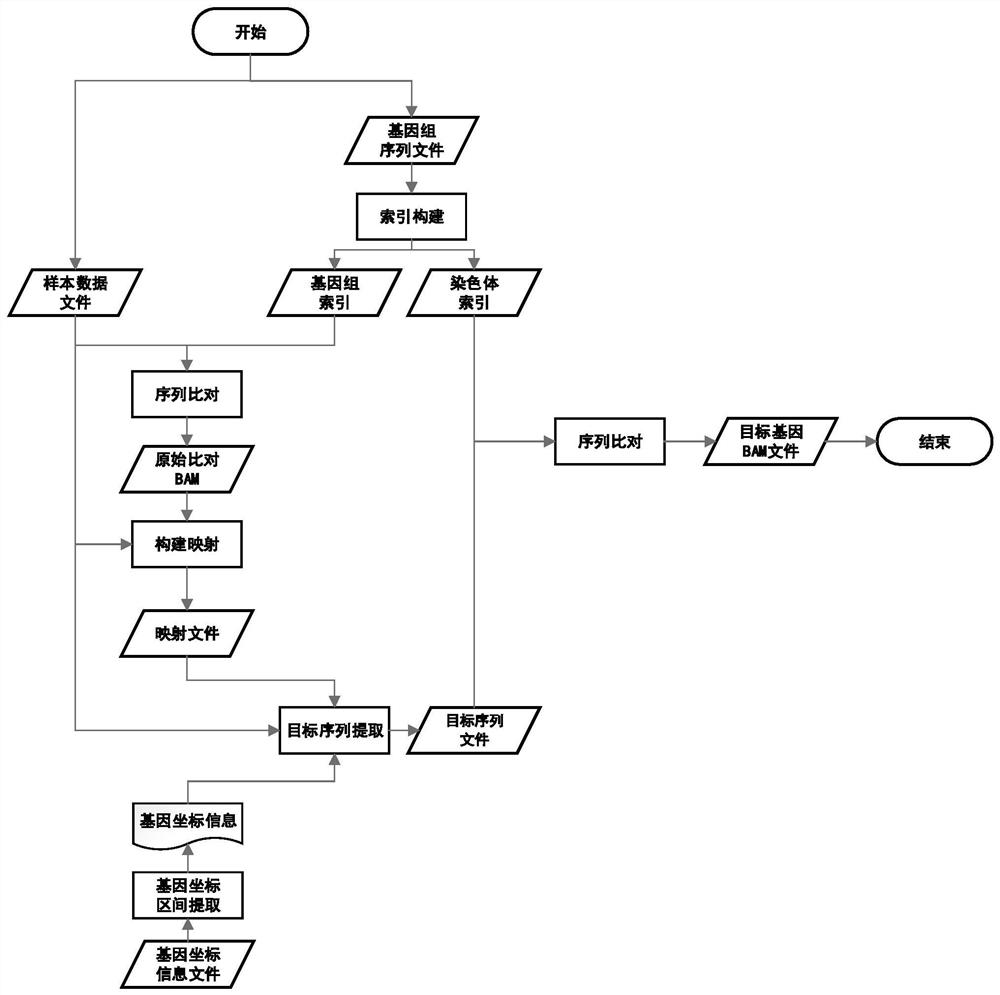
[0049] Example 1 The method of quickly realizing the comparison result data of the target genome region from the raw sequencing data of the sample
[0050] Overview of the overall process:
[0051] (1) Construction of reference genome and chromosome index;
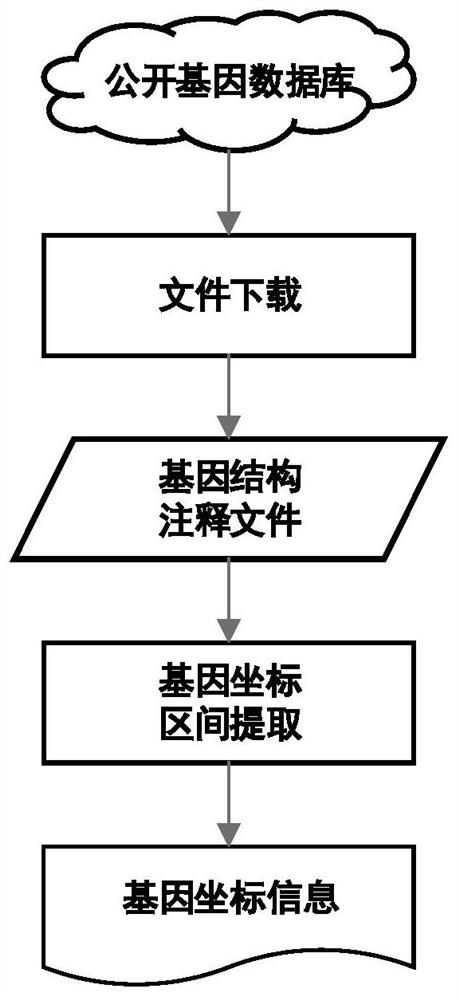
[0052] (2) Acquisition of target gene coordinate interval;
[0053] (3) Construction of mapping files;
[0054] (4) Target sequence file generation;
[0055] (5) Target sequence chromosome alignment and BAM reconstruction.
[0056] Detailed method flow and module explanation:
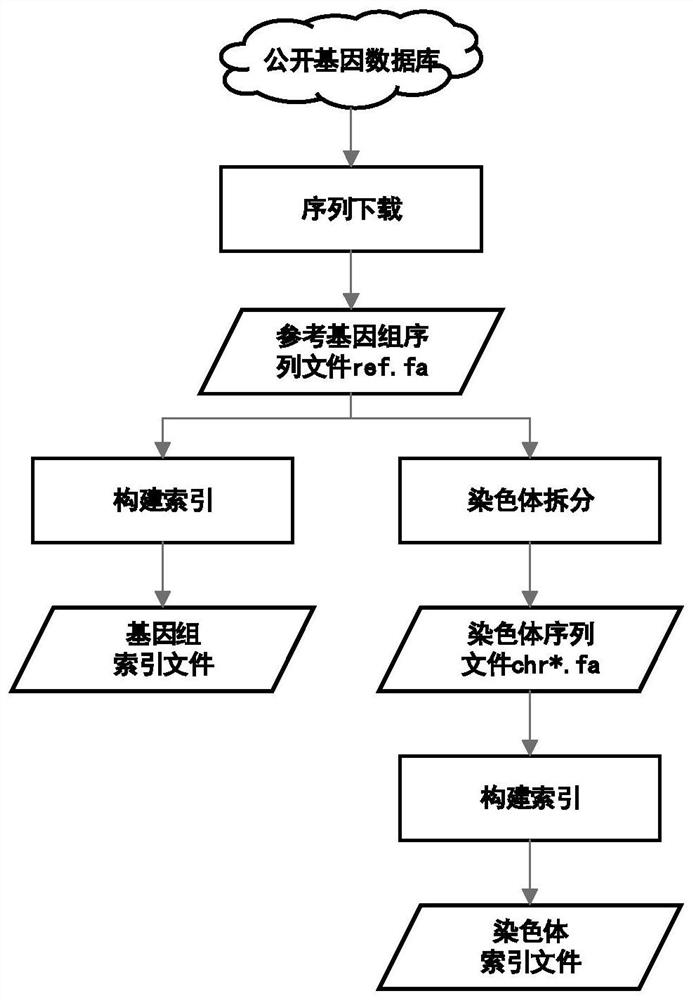
[0057] (1) Construction of reference genome and chromosome index
[0058] Please refer to figure 2 , the function of this step is to build a reference genome index file, which is used to compare the sample data to the reference genome, and obtain the relevant coordinate position information of the related sequence in the genome, and be used for subsequent construction of the mapping file. The construction of the chromosome index is used to q...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com