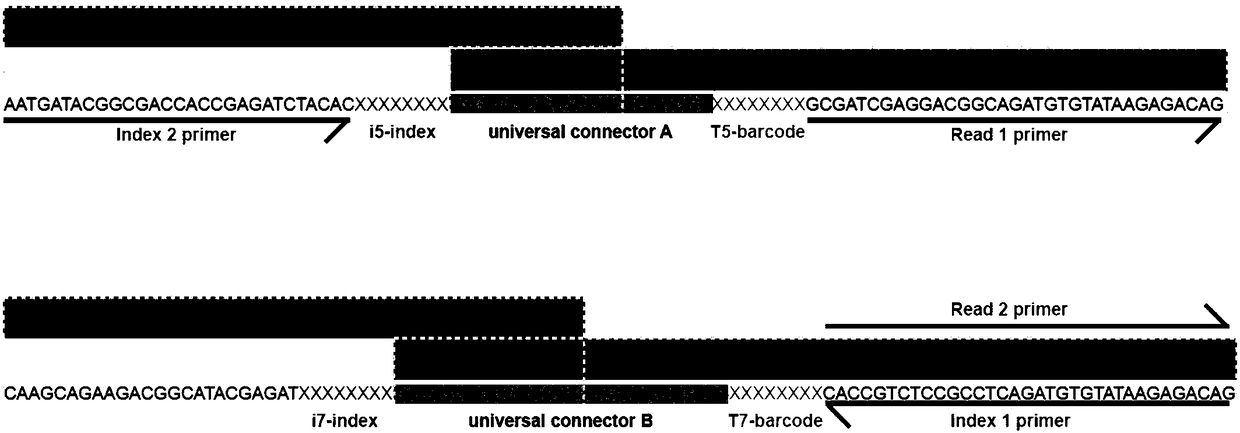
Trace cell ChIP (Chromatin Immunoprecipitation) method
A micro-cell and cell technology, applied in the field of molecular biology experiments, can solve the problems of low detection resolution, unfavorable transcription factor research, information loss, etc., and achieve high efficiency in library construction, improved collection of histone information, and good efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
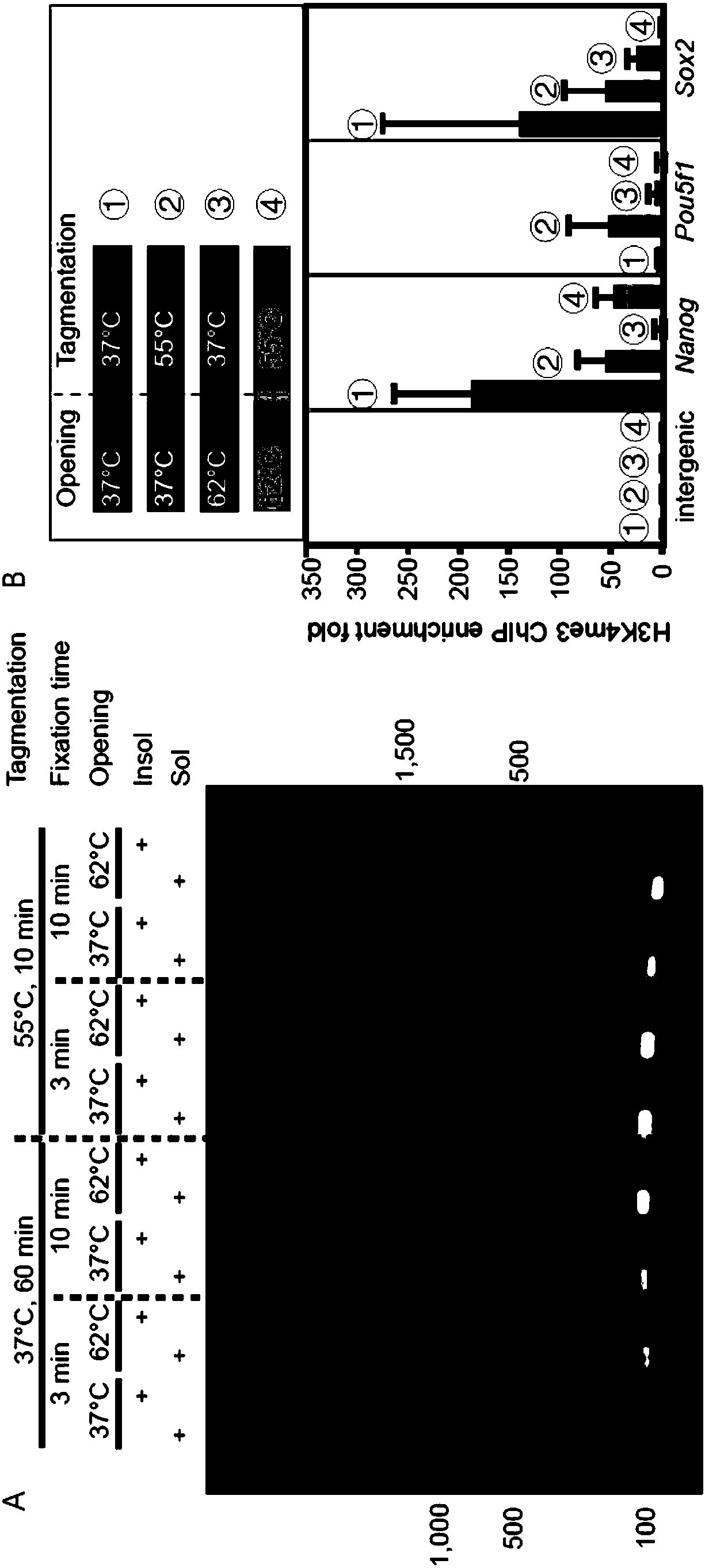
[0091] This embodiment provides a ChIP method for micro cells (1-10000)
[0092] (1) Crosslink the cells with formaldehyde (room temperature) at a concentration of 1v / v% (crosslinking 3min), and collect the cells into 10 μl of hypotonic lysis buffer containing a final concentration of 3w / v% SDS by FACS or mouth picking ( 50mM HEPES (pH 7.9), 30mM KCl, 7v / v% glycerol, 0.3v / v% NP-40, 3w / v% SDS), incubate at 37°C for 3min in PCR;
[0093] (2) Add Triton X-100 to a final concentration of 1v / v% (1v / v% Triton X-100, 0.03w / v% SDS, 15mM EDTA, 200mM NaCl, 30mM Tris-HCl, pH=7.8), after mixing Incubate in a PCR instrument at 37°C for 40min;
[0094] (3) Use Q800R non-contact ultrasonic instrument to further assist in opening the tight structure of chromatin, 160Hz intensity, ultrasonic 15s;
[0095](4) Prepare enzyme digestion system for enzyme digestion reaction: a total of 20 μl, containing all chromatin samples (about 11.2 μl) obtained from microcell processing, 0.1-1 μl Tn5-complex...
Embodiment 2
[0102] Same as Example 1, the only difference is that in step 1), the time for formaldehyde crosslinking (Fixation time) is 10min.
Embodiment 3
[0104] Same as Example 1, the only difference is that in step 1), the temperature of SDS treatment (Opening) is 62°C.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com