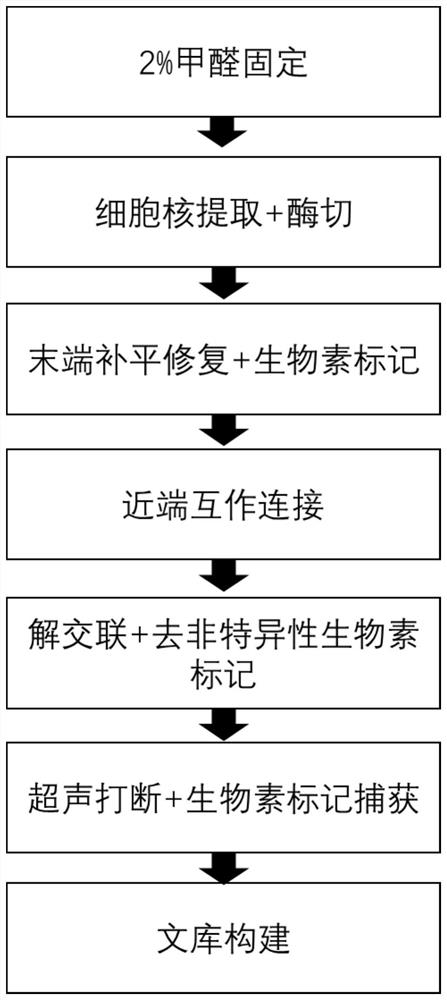
Construction method of cell Hi-C sequencing library
A construction method and sequencing library technology, applied in chemical libraries, library creation, combinatorial chemistry, etc., can solve the problems of low output of library construction, high cost of library construction, and high output of invalid data, so as to improve the efficiency of library construction Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] 1 cell fixation
[0040] (1) After counting cells, resuspend in 750 μL complete cell culture medium, add 41.7 μL 37% formaldehyde (final concentration 2%), and fix at room temperature for 15 minutes (invert once every 1-2 minutes). Remarks: After cell counting, resuspend in 10ml 1x PBS, add 278μL 37% formaldehyde (final concentration 1%), and fix at room temperature for 10 minutes.
[0041] (2) Add 83.25 μL of 2.5M glycine solution, mix gently, let stand at room temperature for 5 minutes, and then place on ice for 15 minutes. Remarks: Add 555 μL of 2.5M glycine solution, mix gently and let stand at room temperature for 5 minutes, then store on ice.
[0042] (3) Centrifuge the sample solution at 400 g for 5 minutes (4° C.), discard the supernatant.
[0043] (4) Resuspend the cells with 500 μL PBS and transfer to a 1.5 mL centrifuge tube. Centrifuge at 400g for 5 minutes at 4°C, discard the supernatant, repeat the operation once, and collect the fixed cells.
[0044] ...
Embodiment 2
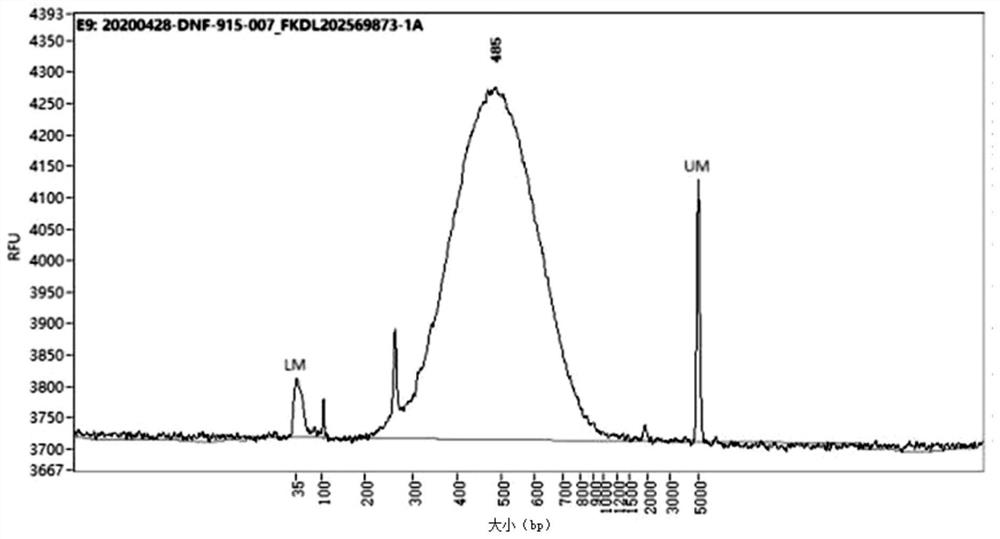
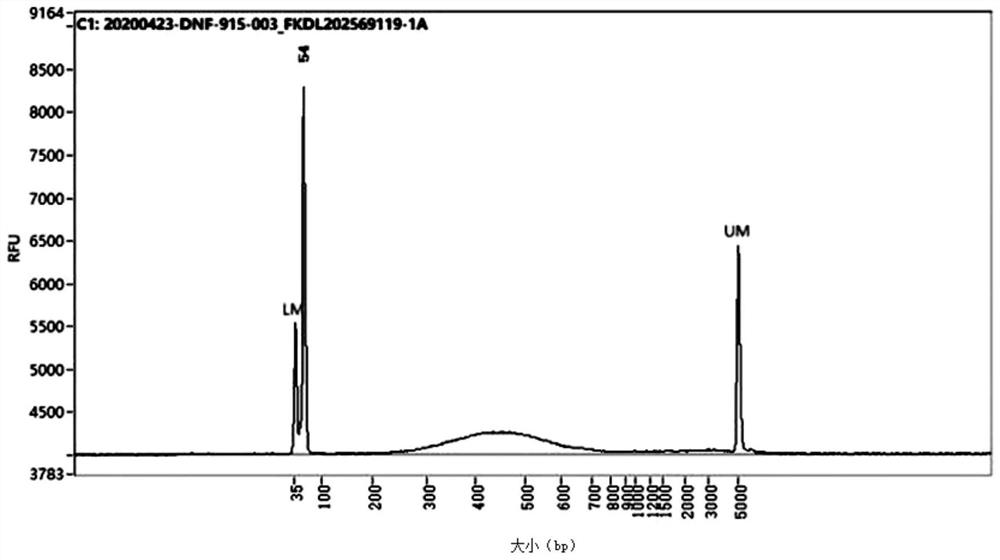
[0134] Utilizing the library construction method under the preferred condition 1 in the present application 1 in Example 1, a Hi-C sequencing library was constructed on duck adipocytes. From Figure 5 and Figure 6It can be seen from the library inspection map of duck adipocytes that the size of the library is concentrated between 400-700bp, which meets the requirements for the distribution of library fragments. The off-machine data of library sequencing was analyzed, and the results are as follows:
[0135] Table 16:
[0136]
[0137] Table 17: Statistics of Duck Adipocyte Sequencing Data
[0138]
[0139] As shown in Table 16 and Table 17, among the data generated from the library constructed by the above 100,000 duck fat cells, the output of invalid data is only 1%, the repetition rate is lower than 20%, and the effective interaction information data is as high as 50%-60% or more.
Embodiment 3
[0141] The comparison of library data between the method of the present invention (100,000 cells) and the conventional cell Hi-C method (5,000,000 cells) is shown in Table 18:
[0142] Table 18:
[0143]
[0144] As shown in Table 18, the Hi-C library data of 100,000 cells using the method of the present invention compared with the library data constructed by the conventional Hi-C method of 5 million cells, the effective interaction information The data reaches more than 60%, and the proportion of invalid data is lower.
[0145] In order to further obtain a better effect of cross-linking and fixation treatment, the inventors also screened the cross-linking and fixation method for cells with a low initial amount, see the following examples for details.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com