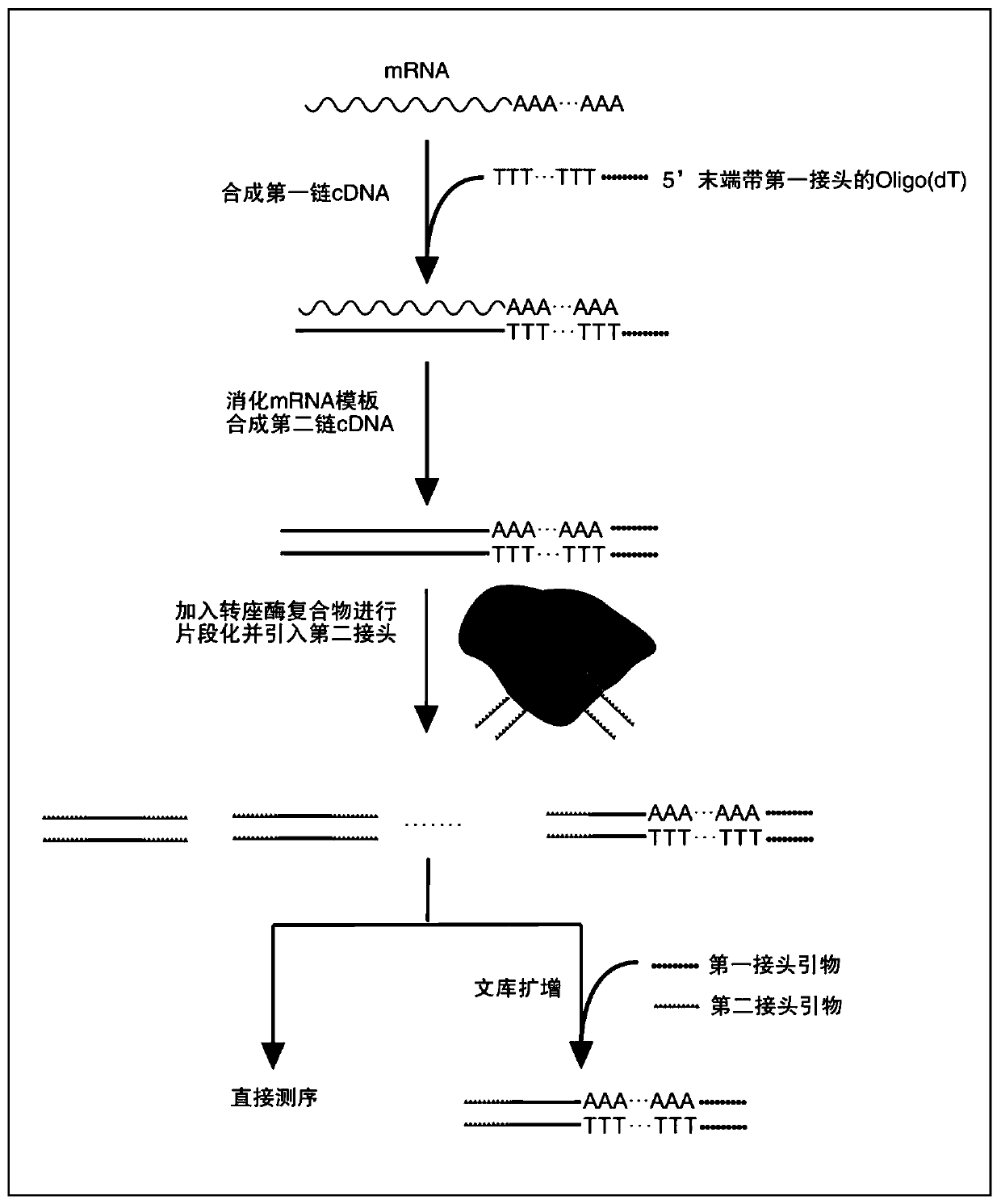
Method for rapid construction of RNA 3' terminal gene expression library
A gene expression and library technology, applied in the field of molecular biology, can solve the problems that the end of the molecule cannot be introduced by a transposase, the sequence may be the same or different, and the two ends of the 3' region of the mRNA molecule cannot be connected by different adapters, so as to avoid information Lost, the method is simple and easy to implement, and the effect of building a library is high
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] Example 1 Entering the double-end sequencing adapter by means of PCR after the transposition method
[0080] 1. Enrichment of mRNA
[0081] For the enrichment of eukaryotic mRNA, the specific steps are conventional mRNA enrichment steps, for example, the mRNA purification kit (Beijing Quanshijin Biology, catalog number: EC801) can be used, and the steps are omitted here.
[0082] 2. Synthesis of first-strand cDNA
[0083] Synthesis of the first-strand cDNA, for example, using the first-strand cDNA transcription synthesis kit (Beijing Quanshijin Biology, catalog number: AT301):
[0084] 1> Add each component according to the following system:
[0085]
[0086] Oligo(T)-VN1 is an Oligo(dT) reverse transcription primer with a first linker, the sequence is: GACGTGTGCTCTTCCGATCTAAAAAAAAAAAAAAAAAAAAAAAAVN (as shown in SEQ ID NO.1), wherein V represents G / A / C among the three bases Any one of the bases, N represents any one of the four bases G / A / C / T.
[0087] 2> Mix gently...
Embodiment 2
[0118] Direct sequencing after embodiment 2 transposition method
[0119] 1. Enrichment of mRNA
[0120] With step 1 in embodiment 1
[0121] 2. Synthesis of first-strand cDNA
[0122] Synthesis of first-strand cDNA was performed. For example, in this example, the first-strand cDNA transcription synthesis kit (Beijing Quanshijin Biology, catalog number: AT301) was used to perform this operation:
[0123] 1> Add each component according to the following system:
[0124]
[0125] Oligo(T)-VN2 is Oligo(dT) reverse transcription primer 2 with a first linker, its sequence is: CAAGCAGAAGACGGCATACGAGAT[index-i5]GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTAAAAAAAAAAAAAAAAAAAAAAAAVN.
[0126] Among them, [index-i5] plays the role of distinguishing the data of each sample, each sample has a different index, and the data of the sample will be distinguished by the difference of the index during sequencing; [index-i5] is inserted into the The original sequence 2 of the Oligo (dT) reverse tra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com