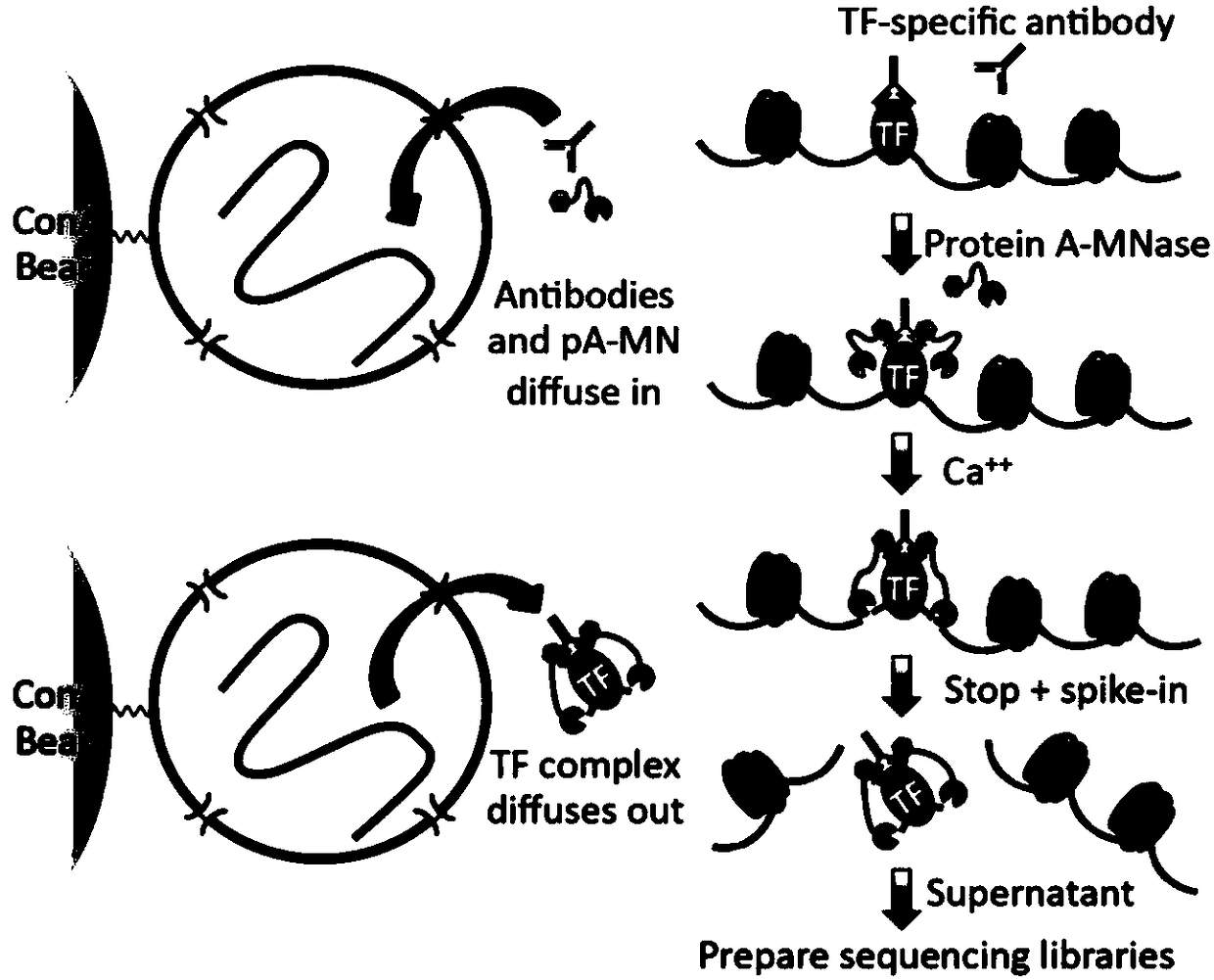
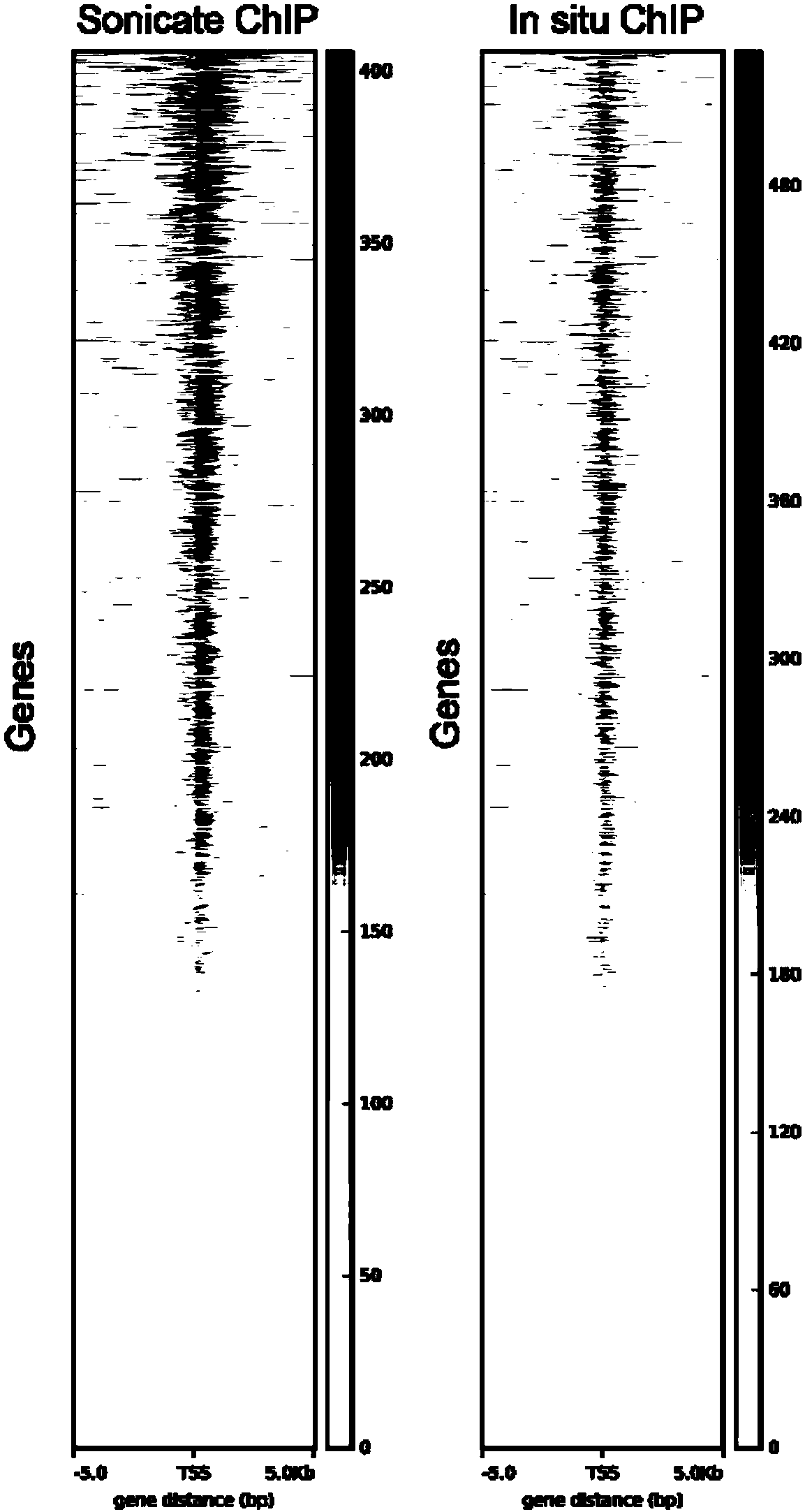
Fusion protein, kit and CHIP-seq detection method
A fusion protein and chip-seq technology, which is applied in the direction of fusion polypeptide, chemical instruments and methods, biochemical equipment and methods, etc., can solve the problems of high probability of experimental failure, high library background, library information loss, etc., and simplify the experimental process , improve the efficiency of database construction, and improve the effect of resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] The present embodiment provides a CHIP-seq detection method (cells are not cross-linked, in a natural state), and the method comprises the following steps:
[0062] 1) Collect about 1,000,000 embryonic stem cells cultured in vitro, wash them twice with PBS, collect the cells by centrifugation, and wash them three times with washing buffer 1;
[0063] 2) Resuspend the cells in binding buffer, add an appropriate amount of antibody, the antibody is H3K4Me3 antibody, and incubate at 4°C for 30 min to fully bind the antibody to the protein;
[0064] 3) Wash the cells 3 times with washing buffer 2 to remove excess unbound antibodies;
[0065] 4) Resuspend the cells with washing buffer 2, then add the fusion protein, and incubate at 4°C for 30 minutes to fully bind the fusion protein to the antibody;
[0066] 5) Wash the cells 3 times with washing buffer 2 to remove excess fusion protein;
[0067] 6) Add the reaction buffer to activate the activity of the fusion protein. In ...
Embodiment 2
[0072] The present embodiment provides a kind of CHIP-seq detection method (cell cross-linking), and described method comprises the following steps:
[0073] 1) Collect 1,000,000 embryonic stem cells, crosslink with 1% FA at room temperature for 3-10min, neutralize glycine and wash with PBS 3 times;
[0074] 2) Resuspend the cells in a hypotonic solution containing 0.3% SDS, and incubate at 37°C for 30 minutes to fully open the chromatin;
[0075] 3) Centrifuge to remove the supernatant;
[0076] 4) Wash cells once with binding buffer, then resuspend cells with binding buffer and add antibody, the antibody is H3K4Me3 antibody, incubate at 4°C for 30 min to fully bind antibody to protein;
[0077] 5) washing the cells with washing buffer 2 for 3 times to remove excess unbound antibodies;
[0078] 6) Resuspend the cells with washing buffer 2, then add the fusion protein, and incubate at 4°C for 30 minutes to fully bind the fusion protein to the antibody;
[0079] 7) washing t...
Embodiment 3
[0085] The present embodiment provides an in situ CHIP-seq detection method, the method comprising the following steps:
[0086] 1) Wash the tissue section 3 times with PBS, and then wash the section 3 times with washing buffer 1;
[0087] 2) Wash the section once with binding buffer, cover the tissue section with binding buffer, add antibody, the antibody is H3K4Me3 antibody, and incubate at 4°C for 1 hour to fully bind the antibody to the protein;
[0088] 3) wash the section with washing buffer 2 for 3 times to remove excess unbound antibody;
[0089] 4) Cover the tissue section with washing buffer 2, then add the fusion protein, and incubate at 4°C for 30 minutes to fully bind the fusion protein to the antibody;
[0090] 5) wash the section with washing buffer 2 for 3 times to remove excess fusion protein;
[0091] 6) Add the reaction buffer to activate the activity of the fusion protein. In order to reduce the reaction background, the reaction is carried out at 4°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com