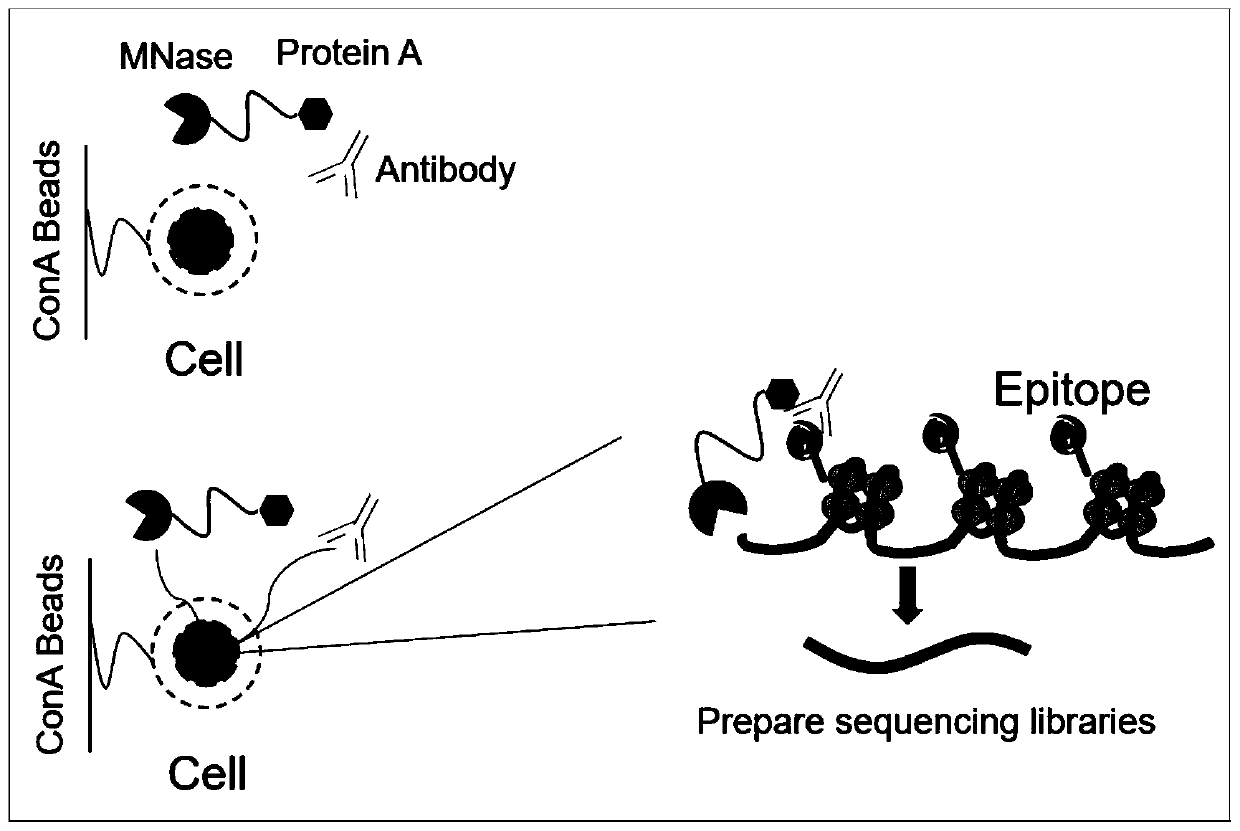
Fusion protein used for single-cell ChIP-seq library preparation and application thereof
A technology of chip-seq and fusion protein, applied in the field of fusion protein prepared by single-cell ChIP-seq library, can solve the problems of library main information loss, high library background, high probability of experimental failure, etc., to reduce background and resolution, reduce Library background, the effect of improving the efficiency of library construction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 5~6
[0062] The fusion protein of Examples 5-6 below is Protein A-Tn5, and the formulations of washing buffer A, washing buffer B, washing buffer C, antibody incubation buffer, etc. used are as follows (for high-throughput single cells):
[0063] Wash buffer A: 20mM HEPES, 150mM NaCl and 0.5mM spermidine;
[0064] Wash buffer B: 20 mM HEPES, 150 mM NaCl, 0.5 mM spermidine, 0.01% (w.t.) digitonin;
[0065] Wash buffer C: 20 mM HEPES, 150 mM NaCl, 0.5 mM spermidine, 0.01% (w.t.) digitonin and 0.1% Triton X-100;
[0066] Activation buffer: 20mM HEPES, 10mM KCl, 1mM CaCl2, 1mM MnCl2
[0067] Antibody incubation buffer: 20mM HEPES, 150mM NaCl, 0.5mM spermidine, 0.01% (w.t.) digitonin, 0.1% Triton X-100 and 2mM EDTA;
[0068] Single-cell reaction buffer: 25 mM Mg2+, 50 mM trismethylaminopropanesulfonic acid and 0.01% (w.t.) digitonin;
[0069] Single cell stop buffer: 40 mM EDTA.
[0070] Single cell sorting buffer: 2% BSA / PBS+2mM EDTA
Embodiment 1
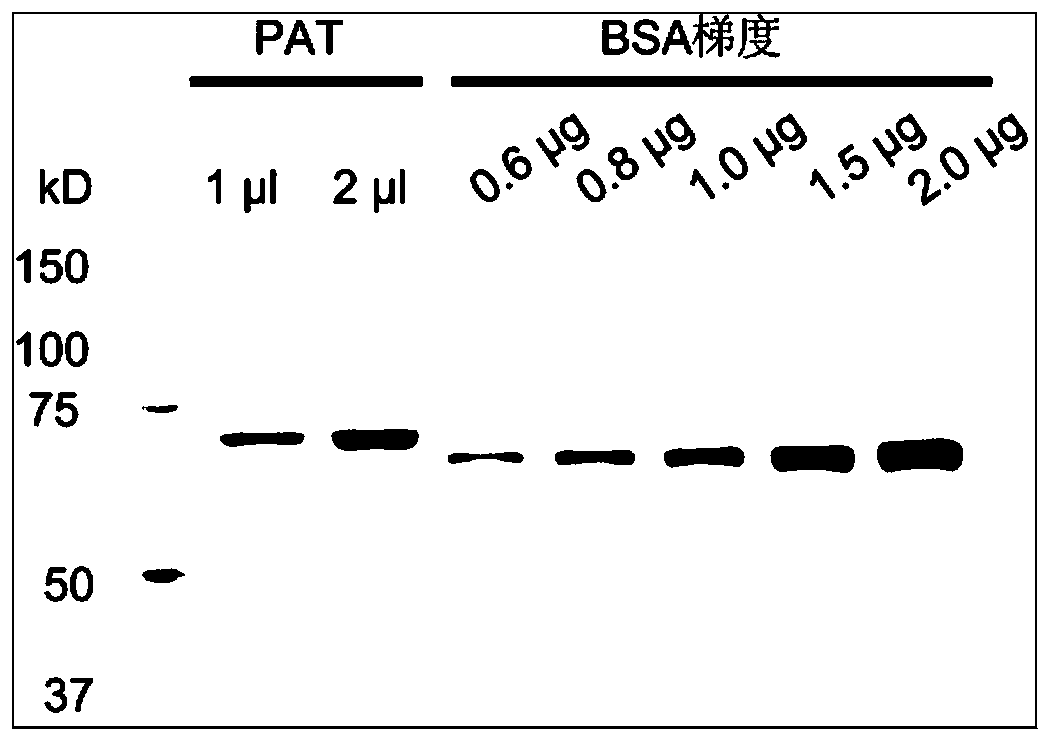
[0072] Embodiment 1, the design, expression and purification of highly active ProteinA-Tn5 (PAT protein)
[0073] This embodiment provides the design and purification of an ultra-high activity Tn5 ((mutant Tn5 enhances enzyme activity) and ProteinA-Tn5 fusion protein. This method has low cost and high protein production efficiency. The method includes the following steps:
[0074] 1. Design of ProteinA-Tn5 fusion protein (PAT)
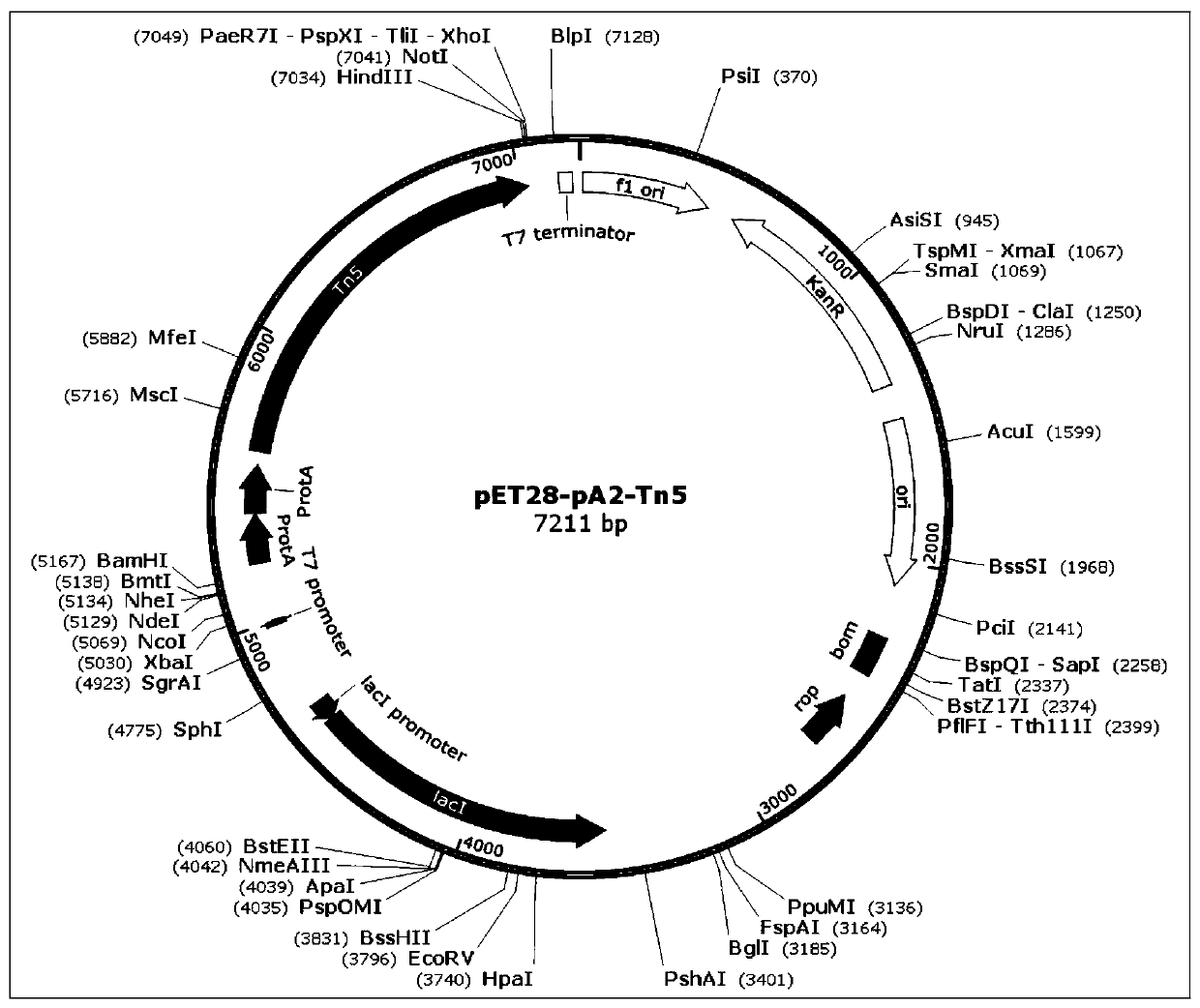
[0075] 1) Cloning the nucleic acid sequence of the mutant Tn5E54K, L372P (hereinafter referred to as Tn5) into the pET28 expression vector, such as figure 2 shown.
[0076] 2) The IgG recognition domain of 2xProtein A is connected.
[0077] 3) Sanger sequencing was used to ensure the authenticity of the coding region.
[0078] Compared with wild-type Tn5, the mutant Tn5 in PAT has stronger adapter binding ability, and its cleavage activity is increased by more than ten times. The improvement of cleavage activity is conducive to its application in v...
Embodiment 2
[0100] Embodiment 2, optimization of each functional block of PAT fusion protein
[0101] 1. Optimal screening of linker sequences
[0102] Selecting a specific linker (-GGSDDDKEF-) sequence to connect different functional fragments of PAT can protect the natural conformation of ProteinA and Tn5, thereby obtaining a fusion protein with better activity.
[0103] The method of connecting fusion proteins with different linkers and the steps of activity verification are as follows:
[0104] 1. When 2xProtein A (IgG recognition sequence) is connected to the original sequence of Tn5, different linker sequences are introduced by PCR (see Table 1, a total of 10 linker sequences).
[0105] Table 1 Different linker sequences
[0106]
[0107]
[0108] 2. Obtain the PAT connected by different linkers according to the method described in Example 1, and then perform functional identification on it, the method is as follows:
[0109] 1) Take 10 μl IgG beads, wash with 200 μl HGX bu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com